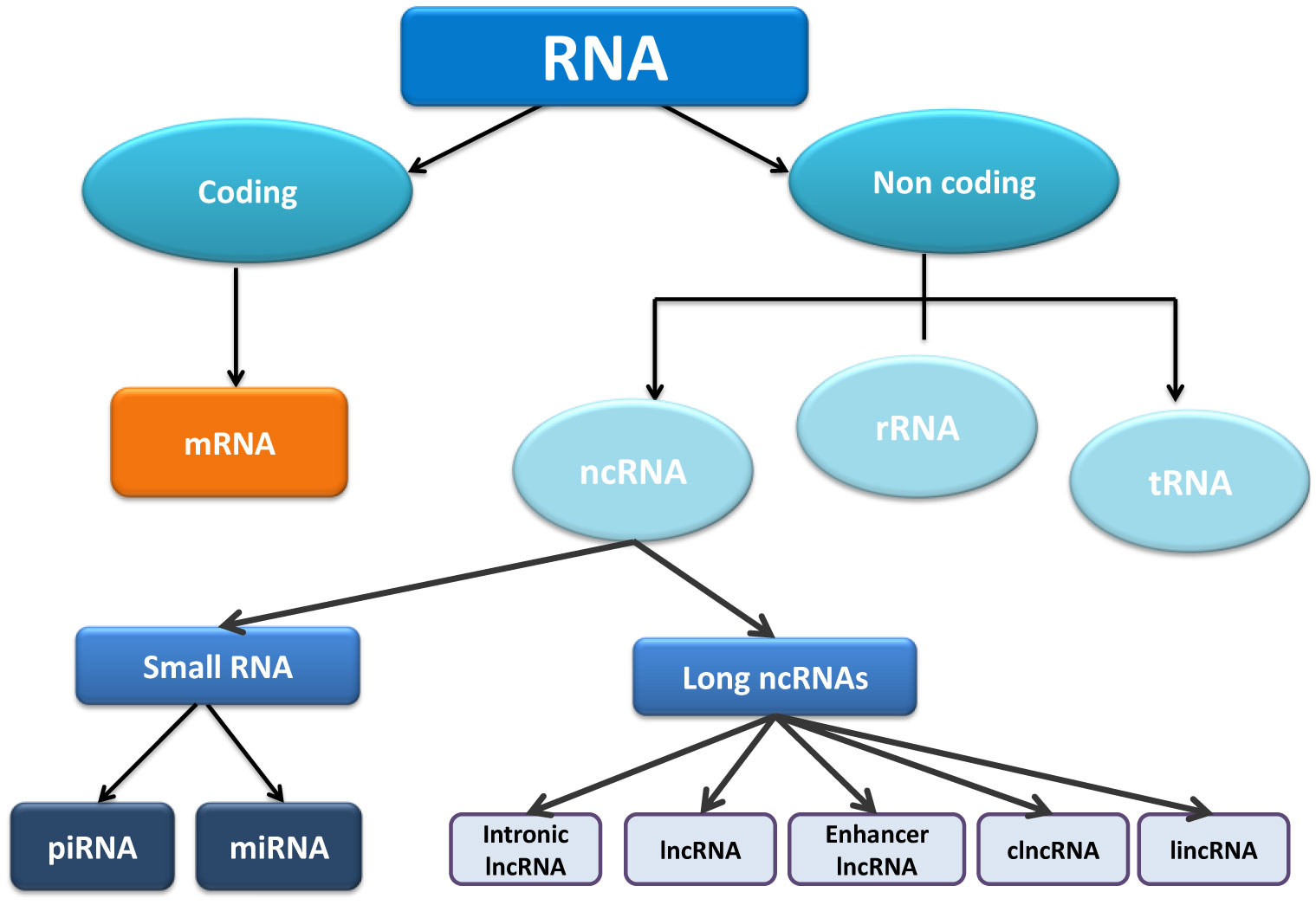
Long non‐coding RNAs (lncRNAs) refer to a class of non‐protein coding RNAs that are more than 0 nucleotides and are differentially accumulated in the nucleus and cytoplasm9 LncRNAs play various regulatory roles in the cell including regulation of development, stem cell pluripotency, cell growth and apoptosis and are frequentlyNONCODEv5, the systematic public database that is dedicated to the collection and annotation of lncRNA, has recorded 548,640 lncRNA transcripts from 17 different species, with the expression profiles of entire human and mouse genome being annotated 6To establish a highquality database of ncRNAs, all the articles related to drug resistance and ncRNAs were manually extracted from publications (Fig 1) (i) We searched all published studies in the PubMed database using the following combination of keywords "long noncoding RNA or lncRNA and drug and cancer," "microRNA or miRNA and drug and cancer," "circRNA and drug and cancer
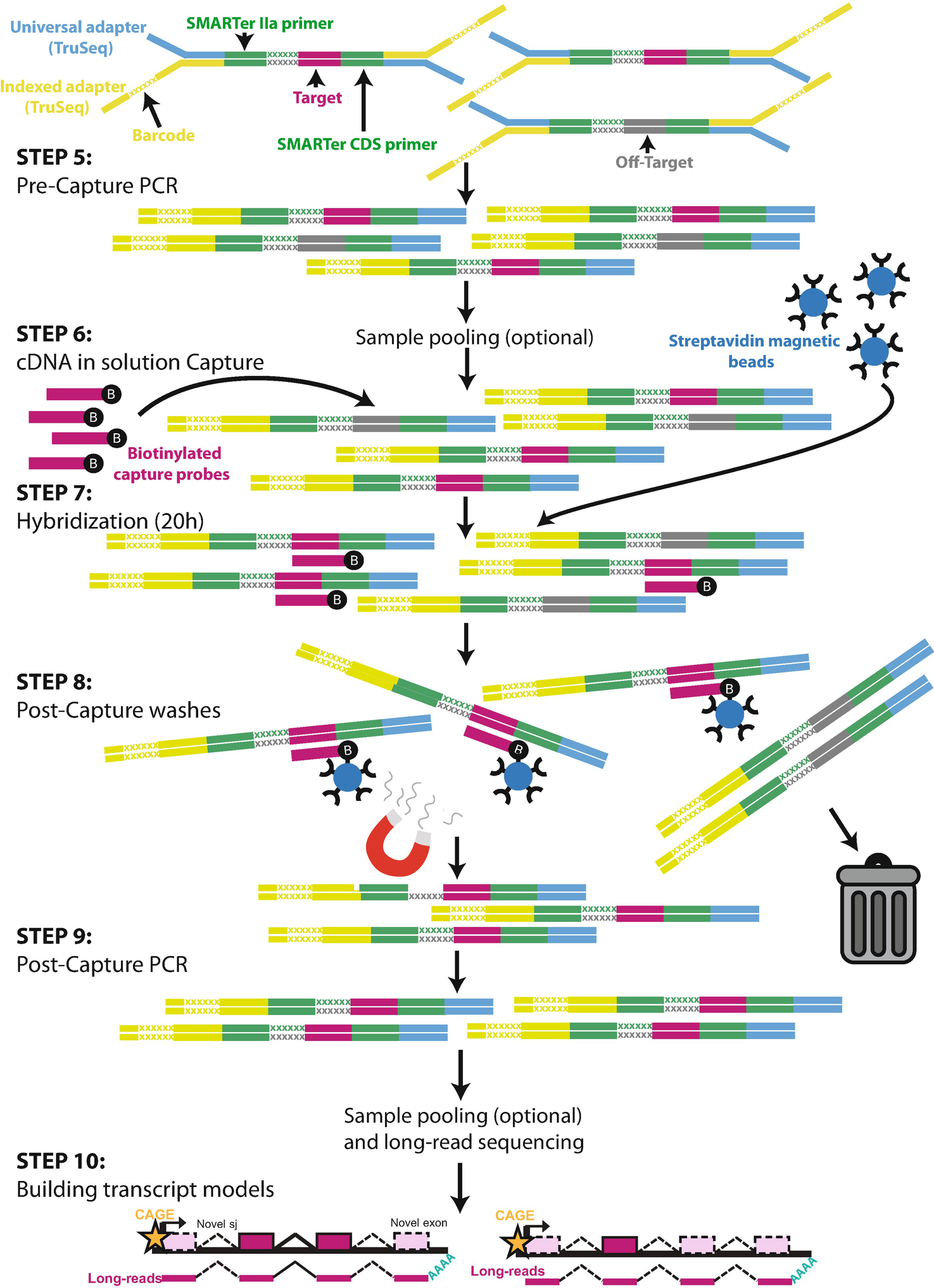
Annotation Of Full Length Long Noncoding Rnas With Capture Long Read Sequencing Cls Springerlink
Noncodev5 a comprehensive annotation database for long non-coding rnas
Noncodev5 a comprehensive annotation database for long non-coding rnas-A comprehensive collection of 270,044 human lncRNAs and systematic curation of lncRNAs' annotation by multiomics data integration, function annotation and disease association MONOCLdb The MOuse NOnCode Lung database provides the annotations and expression profiles of mouse long noncoding RNAs (lncRNAs) involved in influenza and SARSCoV(iii) the RNA secondary structures of
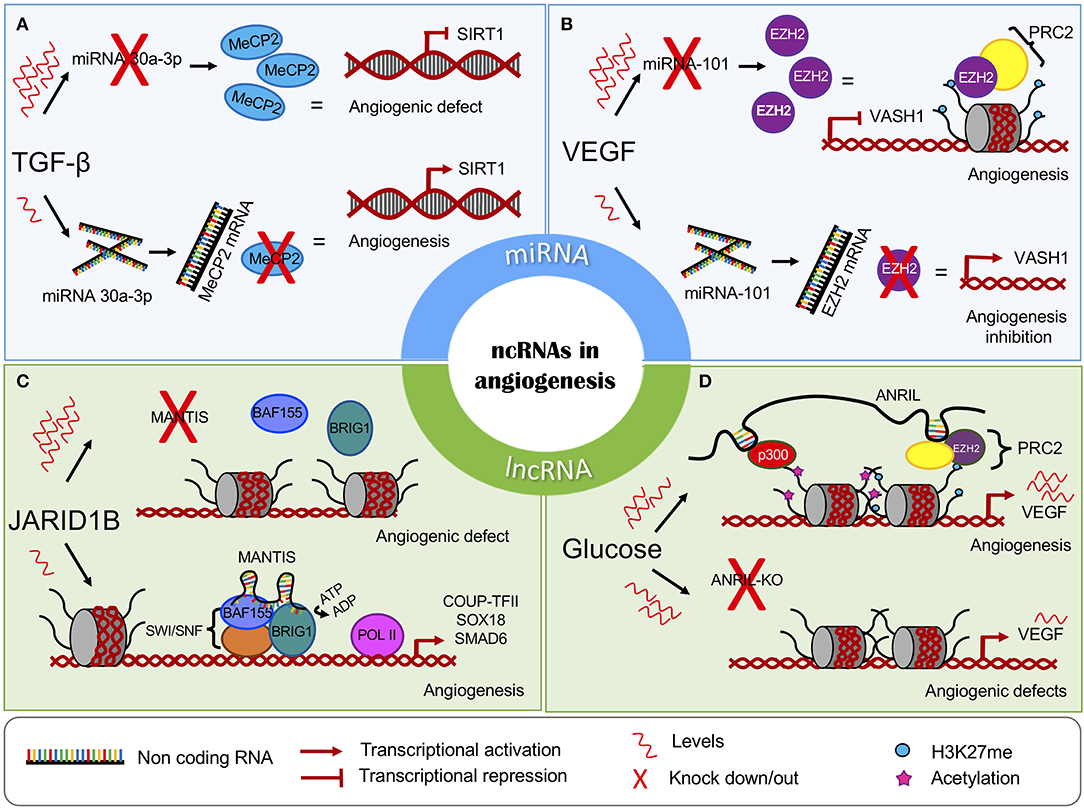


Frontiers The Regulatory Roles Of Non Coding Rnas In Angiogenesis And Neovascularization From An Epigenetic Perspective Oncology
NONCODEV5 a comprehensive annotation database for long noncoding RNAs Nucleic Acids Res 46, D308–D314 doi /nar/gkx1107 PubMed Abstract CrossRef Full Text Google ScholarIntroduction Long noncoding RNAs (lncRNAs) are a recently discovered class of non protein coding transcripts encoded by many metazoan genomes Members of this class have been largely annotated in the recent years following the transcriptome annotation of metazoans using deep sequencing approaches 2–5By definition, lncRNAs are transcripts with a length of more than 0 nucleotides andGTF Long noncoding RNA gene annotation CHR It contains the comprehensive gene annotation of lncRNA genes on the reference
NONCODEV5 – a comprehensive annotation database for long noncoding RNAs November 17, 17 Leave a comment 4,627 Views NONCODE is a systematic database that is dedicated to presenting the most complete collection and annotation of noncoding RNAs (ncRNAs), especially long noncoding RNAs (lncRNAs)Skeletal muscle long noncoding RNAs (lncRNAs) were reported to be involved in the development of type 2 diabetes (T2D) However, little is known about the mechanism of skeletal muscle lncRNAs on hyperglycemia of diabetic GotoKakizaki (GK) rats at the age of 3 and 4 weeks To elucidate this, we used RNAsequencing to profile the skeletal muscle transcriptomes including lncRNAs and mRNAs, inNONCODEV5 – a comprehensive annotation database for long noncoding RNAs November 17, 17 Leave a comment 4,464 Views NONCODE is a systematic database that is dedicated to presenting the most complete collection and annotation of noncoding RNAs (ncRNAs), especially long noncoding RNAs (lncRNAs)
57 Fang S, Zhang L, Guo J, et al NONCODEV5 a comprehensive annotation database for long noncoding RNAs Nucleic Acids Res 18;46D308D314 58 Consortium TEP An integrated encyclopediaNONCODEV5 – a comprehensive annotation database for long noncoding RNAs NONCODE is a systematic database that is dedicated to presenting the most complete collection and annotation of noncoding RNAs (ncRNAs), especially long noncoding RNAs (lncRNAs)NONCODE is a comprehensive database that aims to present the most complete collection and annotation of noncoding RNAs, especially long noncoding RNAs (lncRNA genes), and thus NONCODE is essential to modern biological and medical research Scientists are producing a flood of new data from which ne



Invited Review Long Non Coding Rnas Important Regulators In The Development Function And Disorders Of The Central Nervous System Cuevas Diaz Duran 19 Neuropathology And Applied Neurobiology Wiley Online Library


Integrated Analysis Of Long Non Coding Rnas And Mrna Profiles Reveals Potential Sex Dependent Biomarkers Of Bevacizumab Erlotinib Response In Advanced Lung Cancer
Lianhe Zhao's 8 research works with 167 citations and 3 reads, including HERB a highthroughput experiment and referenceguided database of traditional Chinese medicineThe number of lncRNAs in NONCODEv5 increased from 527 336 to 548 640 NONCODEv5 also introduced three important new features (i) human lncRNAdisease relationships and single nucleotide polymorphismlncRNAdisease relationships were constructed;Data LNCipedia is a public database for long noncoding RNA (lncRNA) sequence and annotation The current release contains 127,802 transcripts and 56,946 genes



Genome Wide Analysis Of Long Non Coding Rnas Responsive To Multiple Nutrient Stresses In Arabidopsis Thaliana Request Pdf
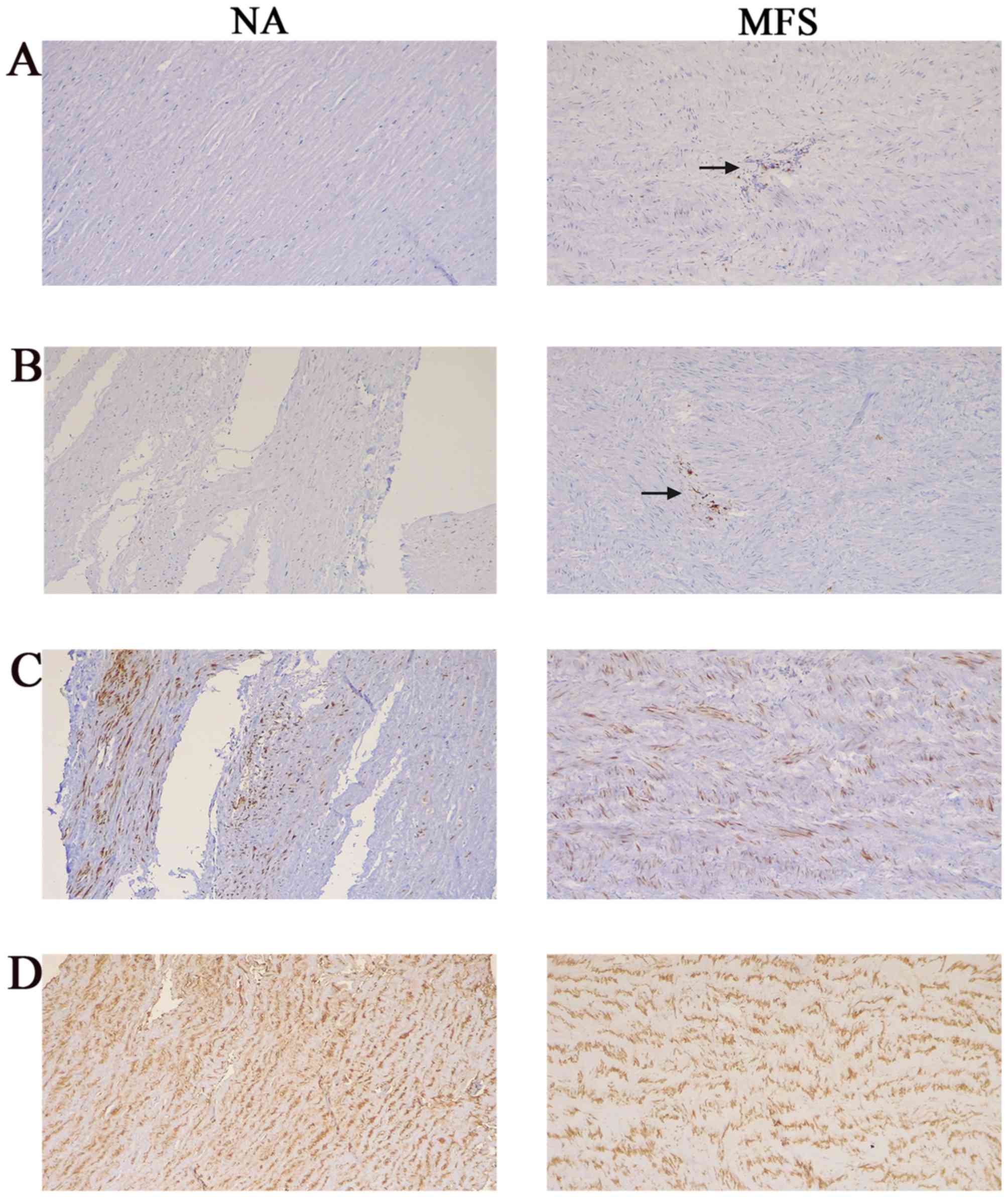


Microarray Analysis Of Long Non Coding Rna Expression Profiles In Marfan Syndrome
, 17 NONCODEV5 a comprehensive annotation database for long noncoding RNAs Nucleic Acids Res 46 D308 – D314 doi /nar/gkx1107 OpenUrl CrossRefData LNCipedia is a public database for long noncoding RNA (lncRNA) sequence and annotation The current release contains 127,802 transcripts and 56,946 genesNutrient stress is the most important environmental stress that limits plant growth and development Although recent evidence highlights the vital functions of long noncoding RNAs (lncRNA) in response to single nutrient stress in some model plants, a comprehensive investigation of the effect of lncRNAs in response to nutrient stress has not been performed in Arabidopsis thaliana



Lncrnakb A Comprehensive Knowledgebase Of Long Non Coding Rnas Biorxiv


Microarray Analysis Of Long Non Coding Rna Expression Profiles In Marfan Syndrome
NONCODEV5 a comprehensive annotation database for long noncoding RNAs Fang S,Zhang L,Guo J,Niu Y,Wu Y,Li H,Zhao L,Li X,Teng X,Sun X,Sun L,Zhang MQ,Chen R,Zhao Y Nucleic Acids Res 17 View Paper (PubMed) View PublicationNONCODEV5 – a comprehensive annotation database for long noncoding RNAs November 17, 17 Leave a comment 4,507 Views NONCODE is a systematic database that is dedicated to presenting the most complete collection and annotation of noncoding RNAs (ncRNAs), especially long noncoding RNAs (lncRNAs)Comprehensive gene annotation ALL It contains the comprehensive gene annotation on the reference chromosomes, scaffolds, assembly patches and alternate loci (haplotypes) This is a superset of the main annotation file;



Pdf Noncodev5 A Comprehensive Annotation Database For Long Non Coding Rnas Semantic Scholar


Germlncrna A Unique Catalogue Of Long Non Coding Rnas And Associated Regulations In Male Germ Cell Development Lncrna Blog
NONCODEV5 a comprehensive annotation database for long noncoding RNAs Fang S,Zhang L,Guo J,Niu Y,Wu Y,Li H,Zhao L,Li X,Teng X,Sun X,Sun L,Zhang MQ,Chen R,Zhao Y Nucleic Acids Res 17 View Paper (PubMed) View PublicationWe describe an updated comprehensive database, LincSNP 30 Emerging evidences have shown the importance of long noncoding RNAs (lncRNAs) as novel regulators of many physical or pathological processes NONCODEV5 a comprehensive annotation database for long noncoding RNAsLong noncoding RNAs (lncRNAs) play important roles in cancerThey are involved in chromatin remodeling, as well as transcriptional and posttranscriptional regulation, through a variety of chromatinbased mechanisms and via crosstalk with other RNA specieslncRNAs can function as decoys, scaffolds, and enhancer RNAs This review summarizes the characteristics of lncRNAs, including their
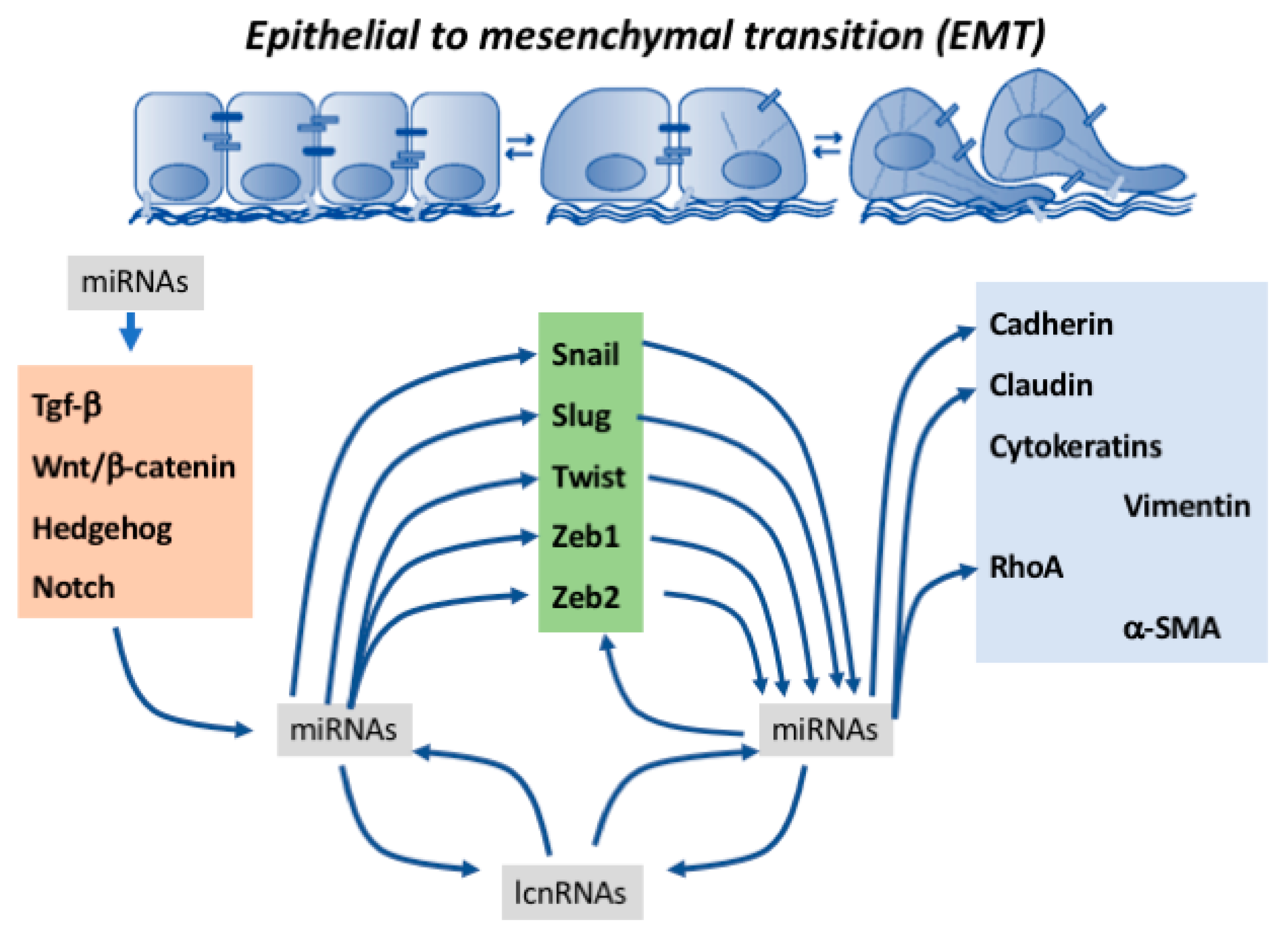


Ncrna Free Full Text Functional Role Of Non Coding Rnas During Epithelial To Mesenchymal Transition Html


Lncrna Expression Profile During Autophagy And Malat1 Function In Macrophages
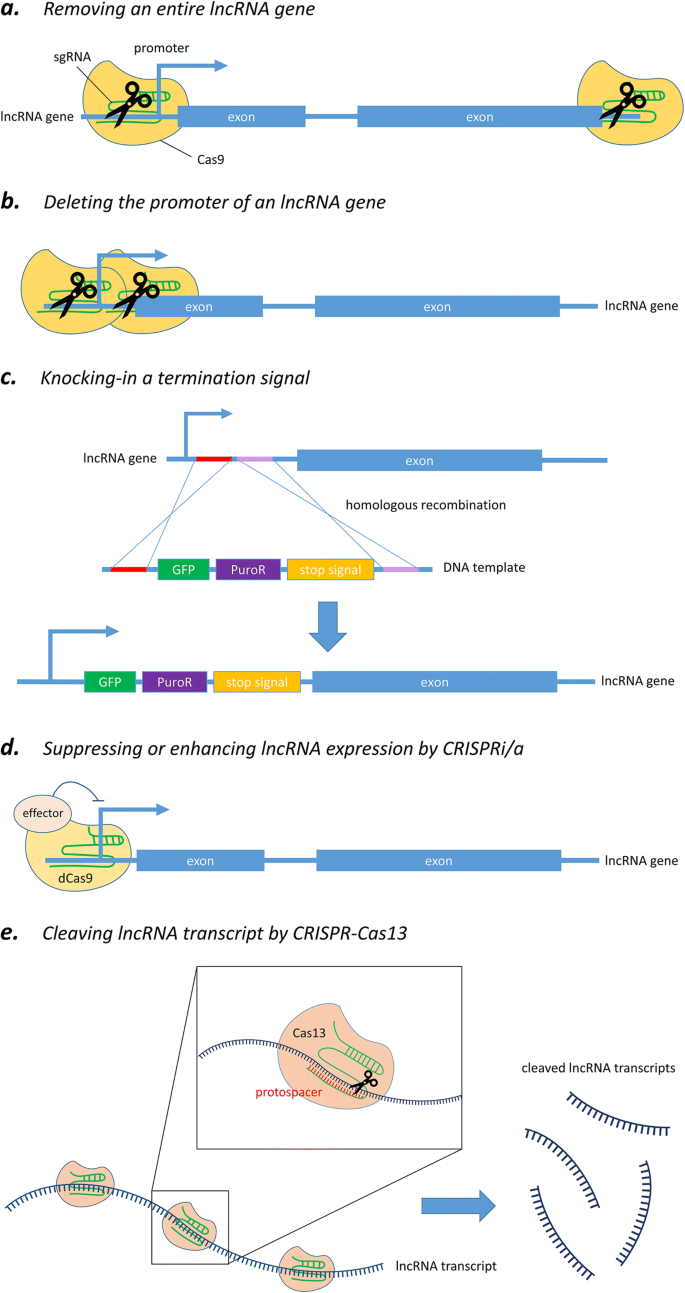
A typical workflow for identification and characterization of novel lncRNAs including the most commonly used methods is depicted in Fig 2Often, novel lncRNA targets are identified by RNAseq experiments either through de novo assembly or derived from an annotation database (eg, GENCODE , LNCipedia , NONCODE or Ensembl)Further characterization of lncRNAs, their isoforms and geneticComprehensive gene annotation ALL It contains the comprehensive gene annotation on the reference chromosomes, scaffolds, assembly patches and alternate loci (haplotypes) This is a superset of the main annotation file;NONCODE (http//wwwbioinfoorg/noncode/) is a systematic database that is dedicated to presenting the most complete collection and annotation of noncoding RNAs (ncRNAs), especially long noncoding RNAs (lncRNAs) Since NONCODE 16 was released two years ago, the amount of novel identified ncRNAs has been enlarged by the reduced cost of nextgeneration sequencing, which has produced an explosion of newly identified data



Molecular Traits Of Long Non Protein Coding Rnas From Diverse Plant Species Show Little Evidence Of Phylogenetic Relationships G3 Genes Genomes Genetics



Invited Review Long Non Coding Rnas Important Regulators In The Development Function And Disorders Of The Central Nervous System Cuevas Diaz Duran 19 Neuropathology And Applied Neurobiology Wiley Online Library
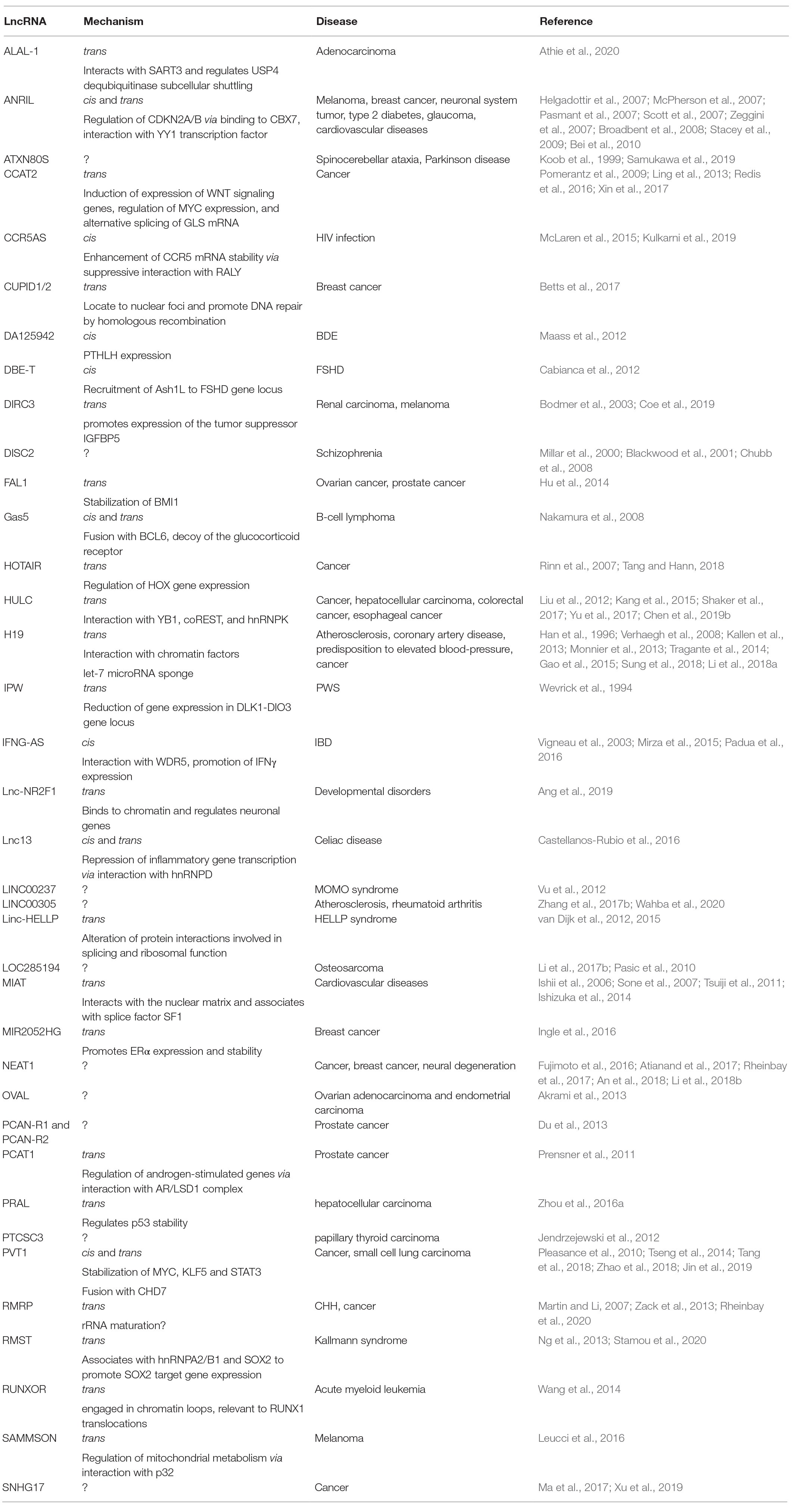
Expression profiles of long noncoding RNAs (lncRNAs) across diverse biological conditions provide significant insights into their biological functions, interacting targets as well as transcriptional reliability However, there lacks a comprehensive resource that systematically characterizes the expAuthors Fang, S, Zhang, L, Guo, J, et al (17) NONCODEV5 a comprehensive annotation database for long noncoding RNAs Nucleic Acids Res Yi Z, Hui L, Fang S, et al NONCODE 16 an informative and valuable data source of long noncoding RNAs J Nucleic Acids Research, 15, 44 (D1)D3D8Abstract Long noncoding RNAs (lncRNAs) are transcripts longer than 0 nucleotides with little or no protein coding potential The expanding list of lncRNAs and accumulating evidence of their functions in plants have necessitated the creation of a comprehensive database for lncRNA research


Annolnc2 The One Stop Portal To Systematically Annotate Novel Lncrnas For Human And Mouse Gao Lab



Computational Methods And Applications For Identifying Disease Associated Lncrnas As Potential Biomarkers And Therapeutic Targets Molecular Therapy Nucleic Acids
Long non‐coding RNAs (lncRNAs) refer to a class of non‐protein coding RNAs that are more than 0 nucleotides and are differentially accumulated in the nucleus and cytoplasm9 LncRNAs play various regulatory roles in the cell including regulation of development, stem cell pluripotency, cell growth and apoptosis and are frequentlyFunctional Analysis of Long NonCoding RNAs Functional Analysis of Long NonCoding RNAs pp Cite as Annotation of FullLength Long Noncoding RNAs with Capture LongRead Sequencing (CLS)Long noncoding RNAs (lncRNAs) are RNAs with a length of over 0 nucleotides that do not have proteincoding abilities Recent studies suggest that lncRNAs are highly involved in physiological functions and diseases lncRNAs HNF1αAS1 and HNF4αAS1 are transcripts of lncRNA genes HNF1αAS1 and HNF4αAS1, which are antisense lncRNA genes located in the neighborhood regions of the
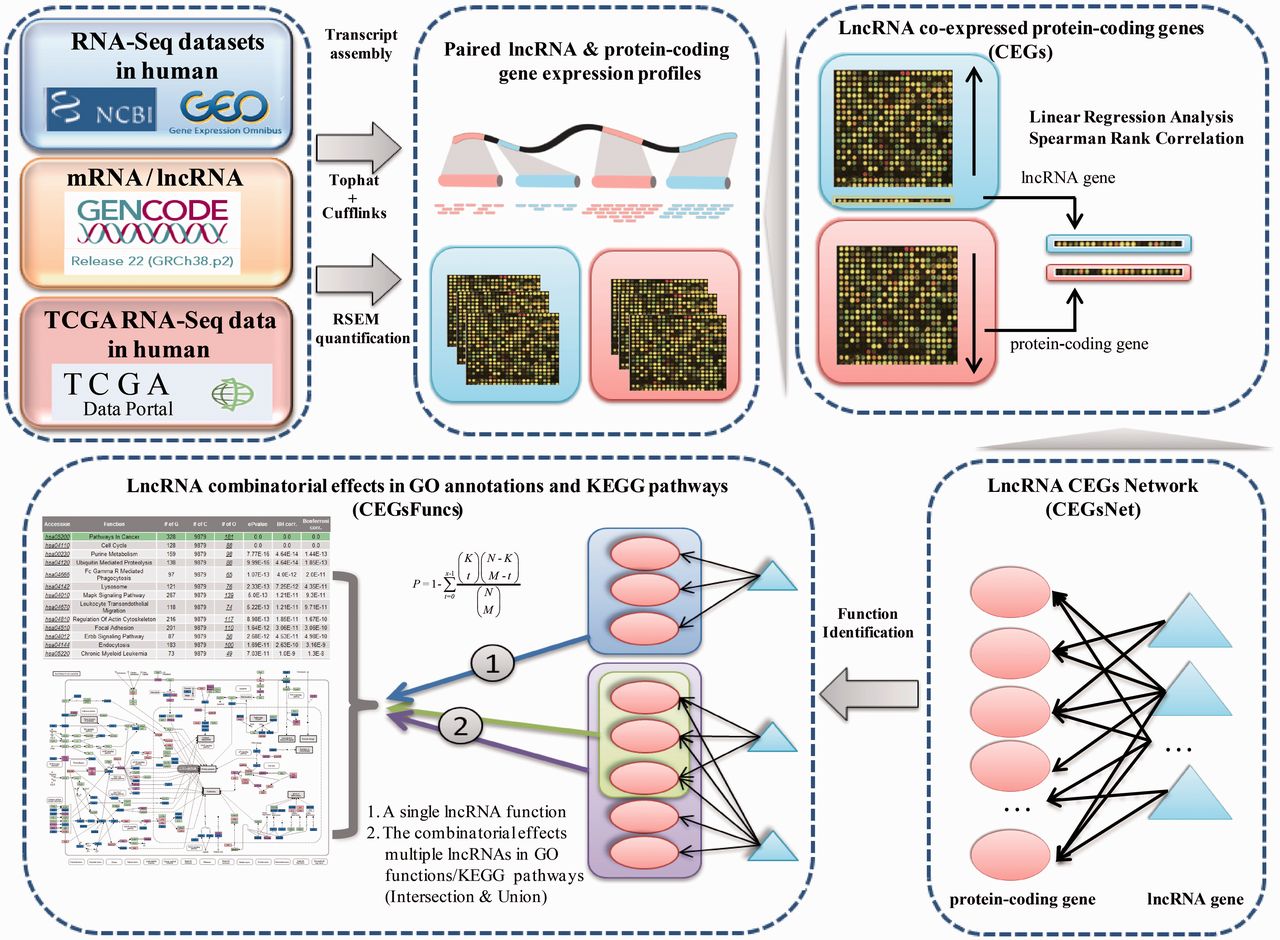


Co Lncrna Investigating The Lncrna Combinatorial Effects In Go Annotations And Kegg Pathways Based On Human Rna Seq Data Lncrna Blog



Pdf Noncode V3 0 Integrative Annotation Of Long Noncoding Rnas Semantic Scholar
Skeletal muscle long noncoding RNAs (lncRNAs) were reported to be involved in the development of type 2 diabetes (T2D) However, little is known about the mechanism of skeletal muscle lncRNAs on hyperglycemia of diabetic GotoKakizaki (GK) rats at the age of 3 and 4 weeks To elucidate this, we used RNAsequencing to profile the skeletal muscle transcriptomes including lncRNAs and mRNAs, inReason is that the functional annotation of long noncoding RNAs (lncRNAs) is largely missing LincRNAs comprise a heterogeneous subset of RNAs that are longer than 0 nucleotides (nt) and are transcribed from intergenic regions without proteincoding potential An increasing number of investigations haveINTRODUCTION There are vast majority of transcribed sequences that do not encode proteins called noncoding RNAs (ncRNAs) Long noncoding RNAs (lncRNAs) are ncRNAs that are >0 nt in length ()Many types of research showed that lncRNAs play key roles in different kinds of biological processes in animals, such as circuitry controlling pluripotency and differentiation, imprinting control
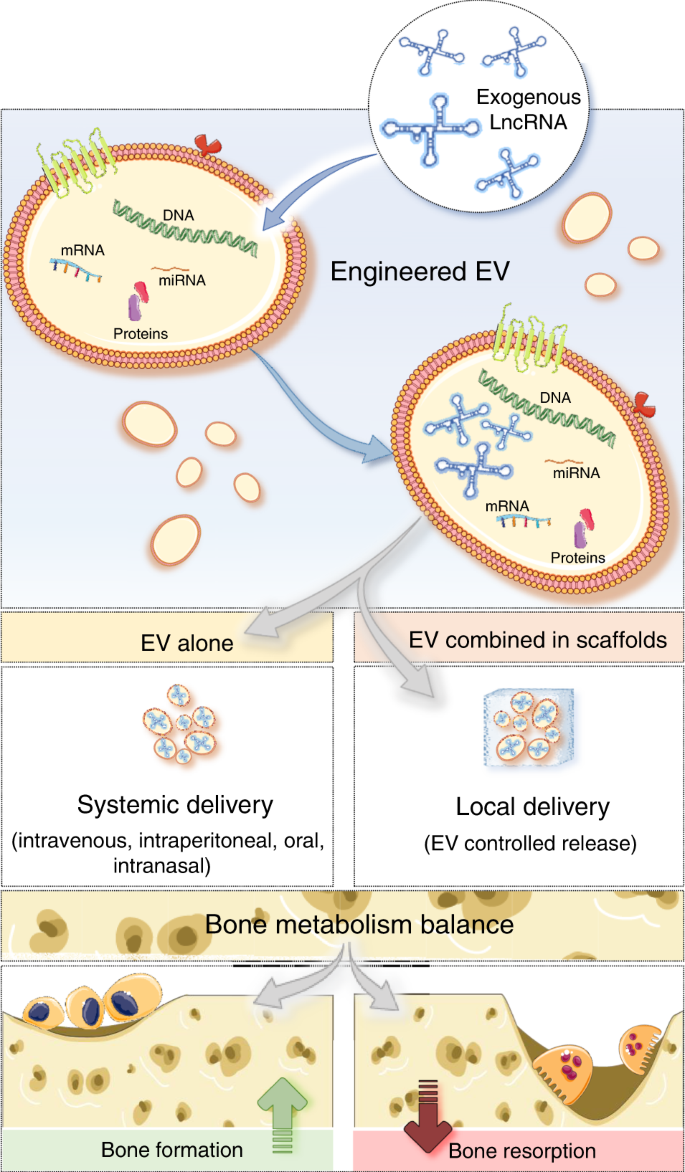


Long Noncoding Rnas A Missing Link In Osteoporosis Bone Research



Lncar A Comprehensive Resource For Lncrnas From Cancer Arrays Cancer Research
GTF Long noncoding RNA gene annotation CHR It contains the comprehensive gene annotation of lncRNA genes on the referenceNONCODEV5 – a comprehensive annotation database for long noncoding RNAs November 17, 17 MNDR v – an updated resource of ncRNAdisease associations in mammalsNONCODE is a comprehensive database that aims to present the most complete collection and annotation of noncoding RNAs, especially long noncoding RNAs (lncRNA genes), and thus NONCODE is



Lncrnakb A Comprehensive Knowledgebase Of Long Non Coding Rnas Biorxiv



Pdf Lncipedia 5 Towards A Reference Set Of Human Long Non Coding Rnas Semantic Scholar
Danshen Transcriptional Resource Database (DsTRD) provides information on transcript sequences and functional annotations including proteincoding RNAs, lncRNAs, other ncRNA, miRNAs and phasiRNAs SoyFN is a resource about soybean miRNA functional networks and functional gene networks It also can be used to seek, download, and analyze the functional networks of soybean miRNAs andThe results bring us closer to a comprehensive annotation of human lncRNAs, though vast amounts of further work are needed to validate the predictions and fully decipher their biology NONCODEV5 a comprehensive annotation database for long noncoding RNAs Nucleic Acids Res (18) SA Mitra et al Several studies have shown that theNONCODE (http//wwwbioinfoorg/noncode/) is an interactive database that aims to present the most complete collection and annotation of noncoding RNAs, especially long noncoding RNAs (lncRNAs) The recently reduced cost of RNA sequencing has produced an explosion of newly identified data
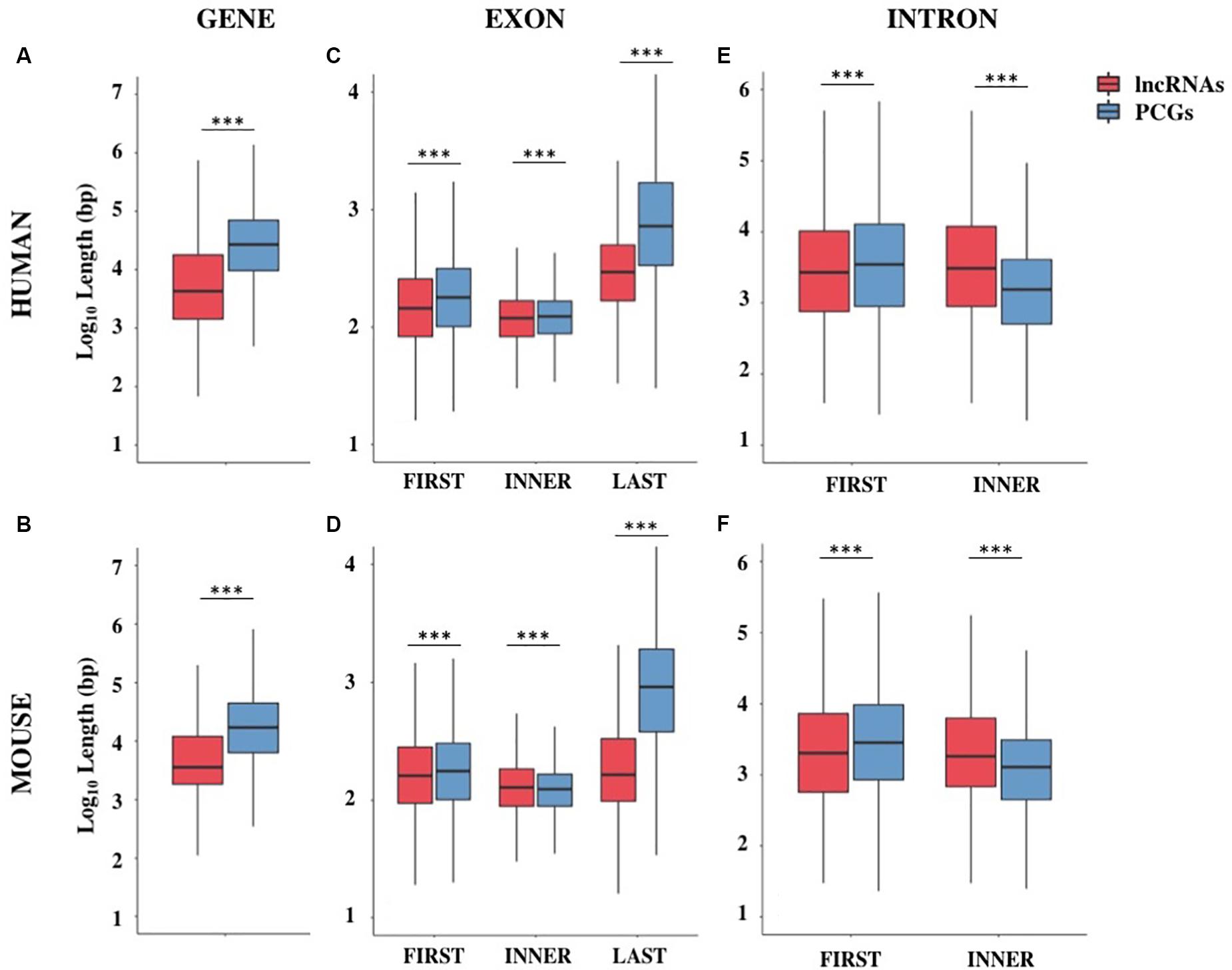


Frontiers Gc Ag Introns Features In Long Non Coding And Protein Coding Genes Suggest Their Role In Gene Expression Regulation Genetics



Annotation Of Full Length Long Noncoding Rnas With Capture Long Read Sequencing Cls Springerlink
NPInter v30 an upgraded database of noncoding RNssociated interactions NPInter v30 an upgraded database of noncoding RNssociated interactions NONCODEV5 a comprehensive annotation database for long noncoding RNAs NONCODEV5 a comprehensive annotation database for long noncoding RNAs FEELnc a tool for long noncoding RNANoncodev5 a comprehensive annotation database for long noncoding RNAs Nuclear Acids Res 46, D308–D314 doi /nar/gkx1107 PubMed Abstract CrossRef Full Text Google ScholarNONCODEV5 a comprehensive annotation database for long noncoding RNAs Nucleic Acids Res 18;
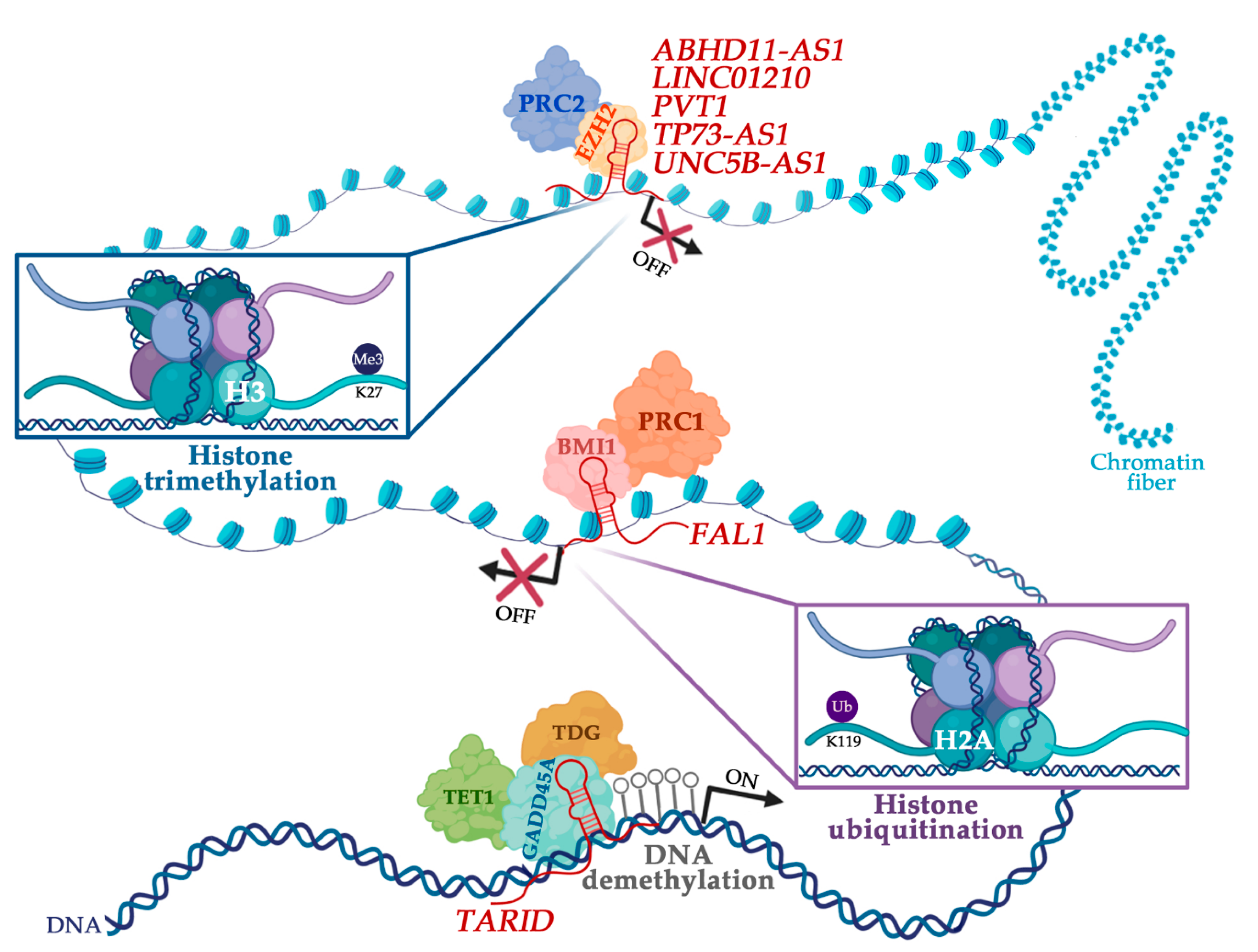


Cancers Free Full Text The Interplay Between Long Noncoding Rnas And Proteins Of The Epigenetic Machinery In Ovarian Cancer Html



Towards A Complete Map Of The Human Long Non Coding Rna Transcriptome Abstract Europe Pmc
NONCODE (http//wwwbioinfoorg/noncode/) is a systematic database that is dedicated to presenting the most complete collection and annotation of noncoding RNAs (ncRNAs), especially long noncoding RNAs (lncRNAs) Since NONCODE 16 was released two years ago, the amount of novel identified ncRNAs has been enlarged by the reduced cost of nextgeneration sequencing, which has produced an explosion of newly identified dataAccumulating evidences have shown that long noncoding RNAs (lncRNAs) can act in cis or trans to perform diverse functions including regulating gene transcription and RNA splicing, modulating the activity or abundance of RNAs and proteins, and organizing nuclear domainsRunsheng Chen's 307 research works with 13,852 citations and 9,533 reads, including NONCODEV6 an updated database dedicated to long noncoding RNA annotation in both animals and plants



Invited Review Long Non Coding Rnas Important Regulators In The Development Function And Disorders Of The Central Nervous System Cuevas Diaz Duran 19 Neuropathology And Applied Neurobiology Wiley Online Library
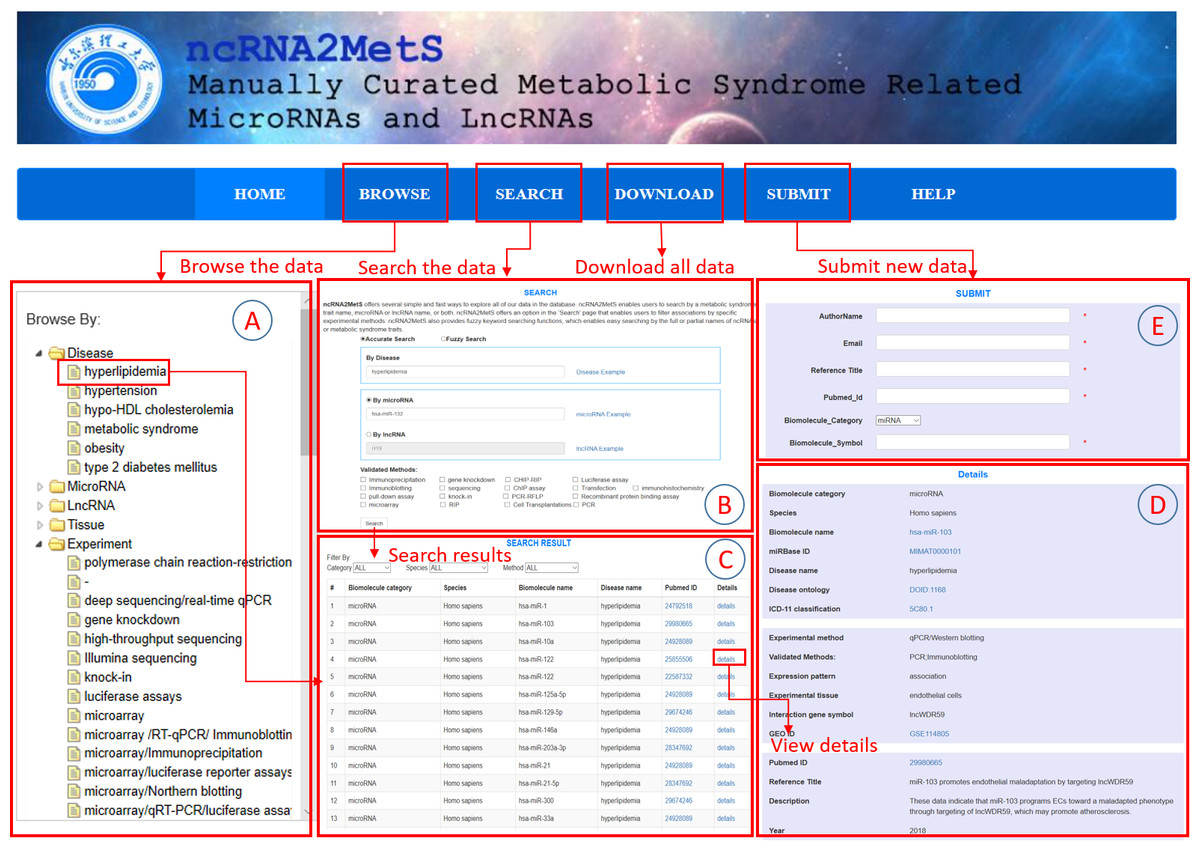


Ncrna2mets A Manually Curated Database For Non Coding Rnas Associated With Metabolic Syndrome Peerj
Fang S, Zhang L, Guo J, Niu Y, Wu Y, Li H, Zhao L, Li X, Teng X, Sun X NONCODEV5 a comprehensive annotation database for long noncoding RNAs Nucleic Acids Res 17;46(D1)D308–14 PubMedCentral CrossRef PubMed Google ScholarMammalian genomes contain tens of thousands of long noncoding RNAs (lncRNAs) that have been implicated in diverse biological processes However, the lncRNA transcriptomes of most mammalian species have not been established, limiting the evolutionary annotation of these novel transcripts Based on RNPInter v30 an upgraded database of noncoding RNssociated interactions NPInter v30 an upgraded database of noncoding RNssociated interactions NONCODEV5 a comprehensive annotation database for long noncoding RNAs NONCODEV5 a comprehensive annotation database for long noncoding RNAs FEELnc a tool for long noncoding RNA
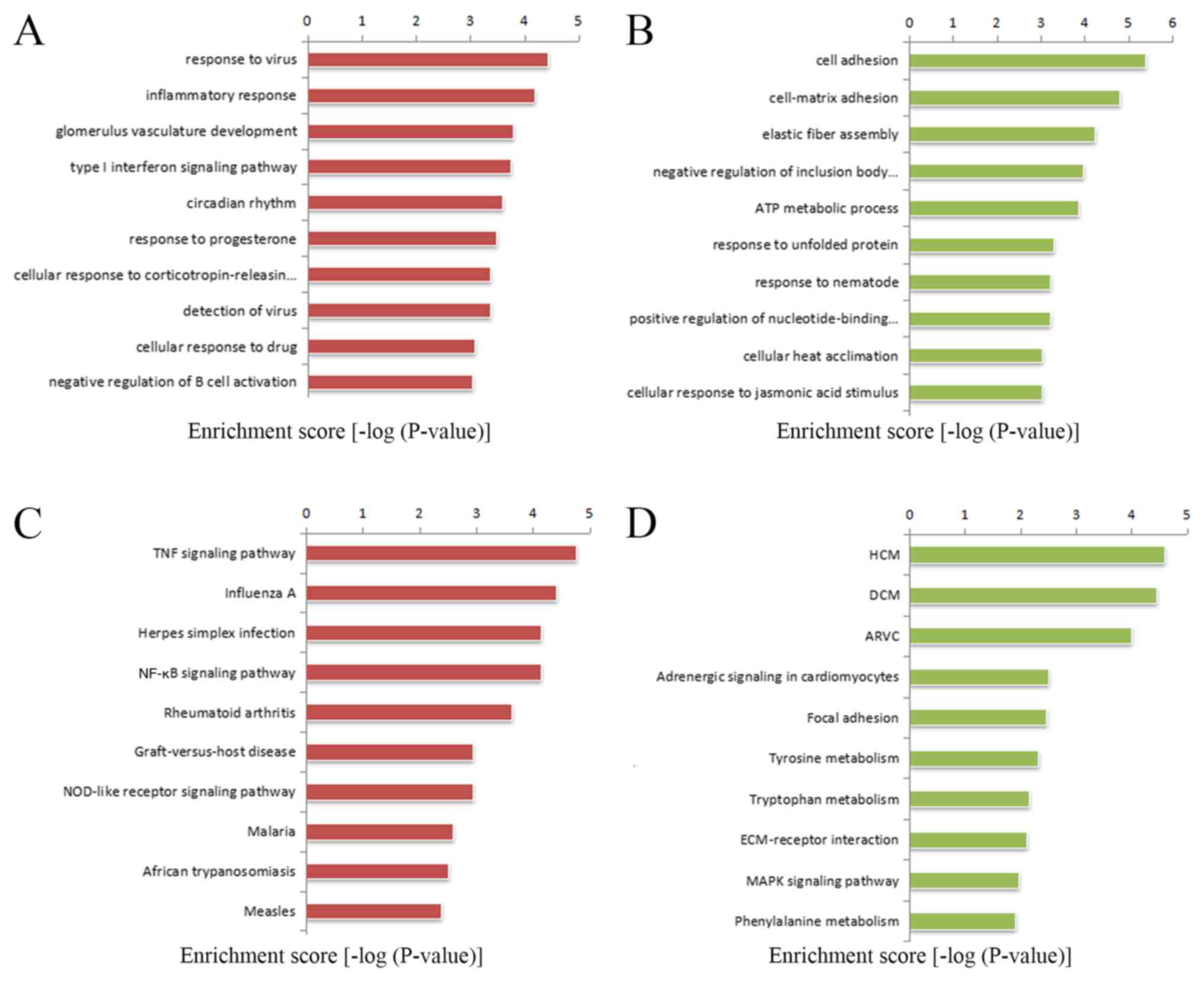


Microarray Analysis Of Long Non Coding Rna Expression Profiles In Marfan Syndrome



Pdf Noncodev5 A Comprehensive Annotation Database For Long Non Coding Rnas
Lnc2Cancer v updated database of experimentally supported long noncoding RNAs in human cancers Nucleic Acids Res 19 Jan 8;47(D1)D1028D1033 doi /nar/gky1096(ii) human exosome lncRNA expression profiles were displayed;NONCODE is a comprehensive database that aims to present the most complete collection and annotation of noncoding RNAs, especially long noncoding RNAs (lncRNA genes), and thus NONCODE is essential to modern biological and medical research Scientists are producing a flood of new data from which ne



Noncodev5 A Comprehensive Annotation Database For Long Non Coding Rnas Lncrna Blog



Phytochemicals As Modulators Of Long Non Coding Rnas And Inhibitors Of Cancer Related Carbonic Anhydrases Abstract Europe Pmc
NONCODEV5 – a comprehensive annotation database for long noncoding RNAs November 17, 17 MNDR v – an updated resource of ncRNAdisease associations in mammalsNONCODE (current version v60) is an integrated knowledge database dedicated to noncoding RNAs (excluding tRNAs and rRNAs) NONCODE v5 is here MoreAndrogen receptor (AR)RNA complexes have been implicated in cancer, including melanoma Schmidt et al demonstrate that AR binds a single strand sequence in the long noncoding RNA (lncRNA) SLNCR Point mutations or oligonucleotides that abrogate AR binding to SLNCR block melanoma invasion, suggesting that targeting lncRNAprotein complexes holds therapeutic promise
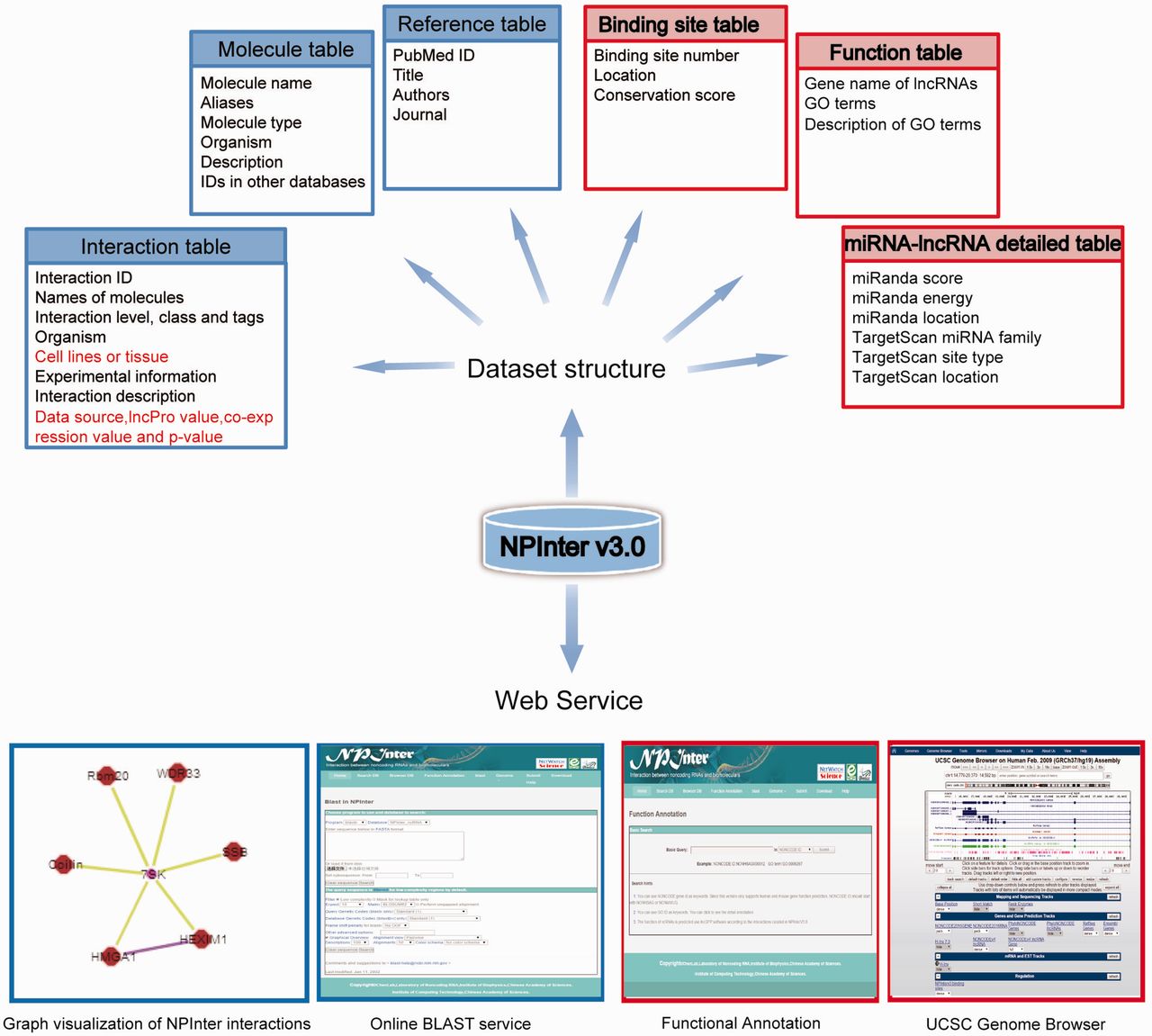


Npinter V3 0 An Upgraded Database Of Noncoding Rna Associated Interactions Lncrna Blog



Lncar A Comprehensive Resource For Lncrnas From Cancer Arrays Cancer Research
Data LNCipedia is a public database for long noncoding RNA (lncRNA) sequence and annotation The current release contains 127,802 transcripts and 56,946 genesBackground Long noncoding RNAs (lncRNAs) are transcripts that are 0 bp or longer, do not encode proteins, and potentially play important roles in eukaryotic gene regulation However, the number, characteristics and expression inheritance pattern of lncRNAs in maize are still largely unknown Results By exploiting available public EST databases, maize whole genome sequence annotation andExpression profiles of long noncoding RNAs (lncRNAs) across diverse biological conditions provide significant insights into their biological functions, interacting targets as well as transcriptional reliability However, there lacks a comprehensive resource that systematically characterizes the expression landscape of human lncRNAs by integrating their expression profiles across a wide range



Frontiers The Regulatory Roles Of Non Coding Rnas In Angiogenesis And Neovascularization From An Epigenetic Perspective Oncology
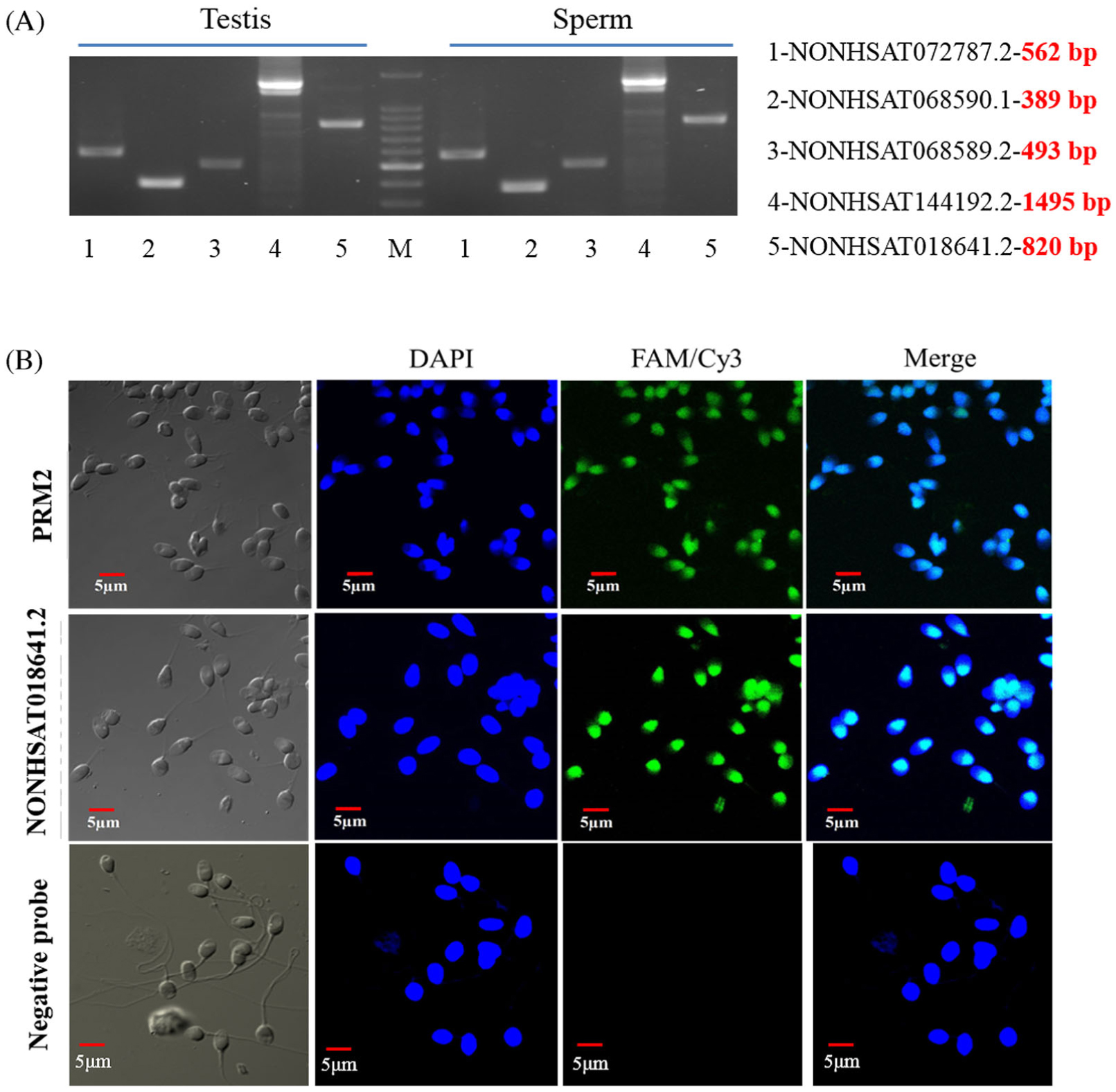


Expression Profiles And Characteristics Of Human Lncrna In Normal And Asthenozoospermia Sperm
Fang S, et al NONCODEV5 a comprehensive annotation database for long noncoding RNAs Nucleic Acids Res 18;46(Database issue)D308–14 Article Google ScholarDuring nuclear maturation of most eukaryotic premessenger RNAs and long noncoding RNAs, introns are removed through the process of RNA splicing Different classes of introns are excised by the U2type or the U12type spliceosomes, large complexes of small nuclear ribonucleoprotein particles and asA team led by researchers at the National Heart, Lung, and Blood Institute has assembled the most comprehensive long noncoding RNA knowledgebase (lncRNAKB) by methodically integrating widely used lncRNAs resources It presents the largest annotation of 77,199 human lncRNAs (224,286 transcripts) including the latest CHESS, FANTOM, LNCipedia, and NONCOD resources



Pdf Community Curation And Expert Curation Of Human Long Noncoding Rnas With Lncrnawiki And Lncbook


Lncrnakb Methods
A comprehensive database that reveals lncRNA expression and variation among different cell groups is still lacking Table 1 List of selected scRNASeq databases NONCODEV5 a comprehensive annotation database for long noncoding RNAs Singlecell analysis of long noncoding RNAs in the developing human neocortexA comprehensive collection of 270,044 human lncRNAs and systematic curation of lncRNAs' annotation by multiomics data integration, function annotation and disease association MONOCLdb The MOuse NOnCode Lung database provides the annotations and expression profiles of mouse long noncoding RNAs (lncRNAs) involved in influenza and SARSCoVNONCODEV5 a comprehensive annotation database for long noncoding RNAs Fang S,Zhang L,Guo J,Niu Y,Wu Y,Li H,Zhao L,Li X,Teng X,Sun X,Sun L,Zhang MQ,Chen R,Zhao Y Nucleic Acids Res 17 View Paper (PubMed) View Publication
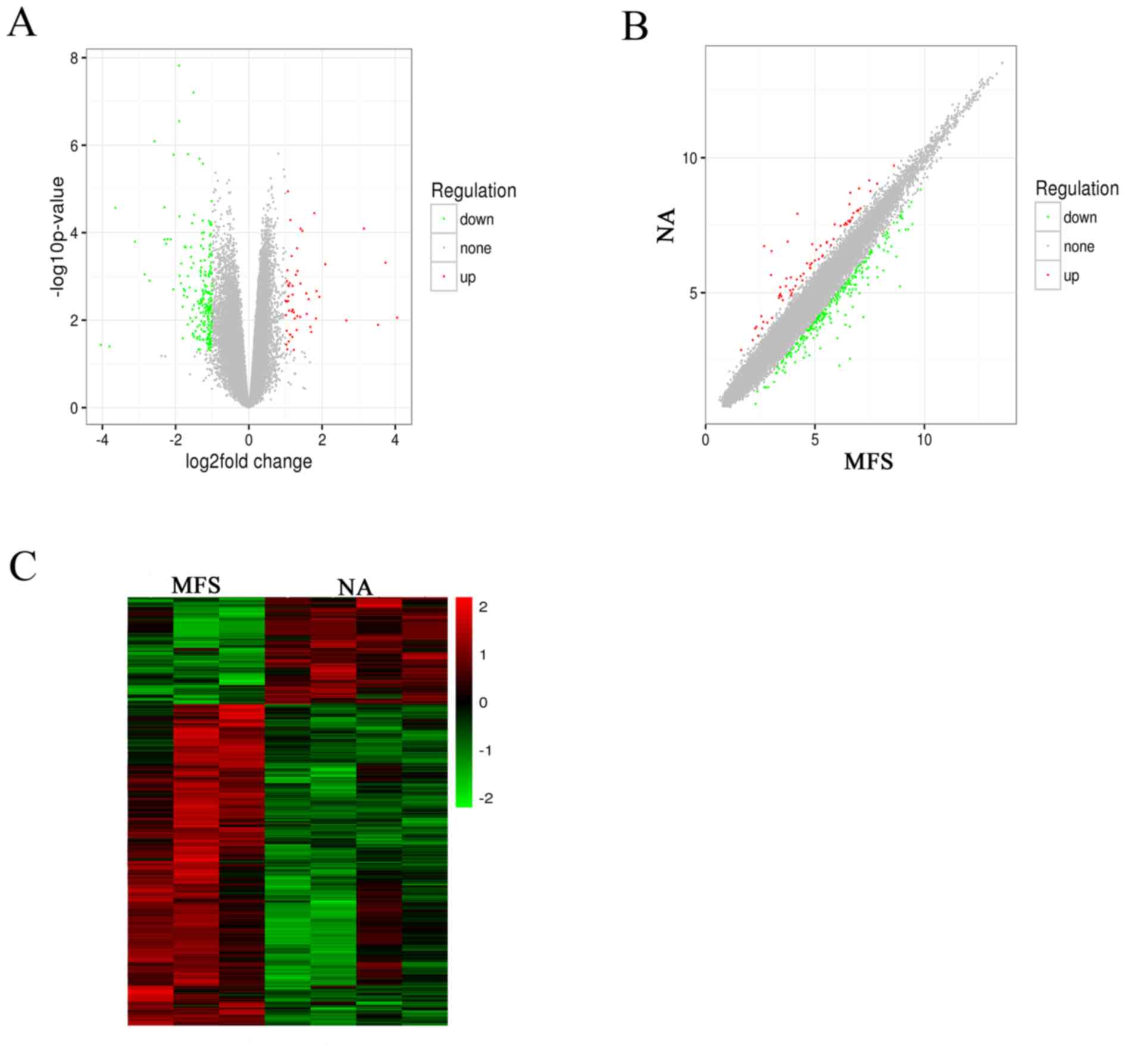


Microarray Analysis Of Long Non Coding Rna Expression Profiles In Marfan Syndrome



Non Coding Rnas Long Non Coding Rnas And Micrornas In Endocrine Related Cancers In Endocrine Related Cancer Volume 25 Issue 4 18
Functional Analysis of Long NonCoding RNAs Functional Analysis of Long NonCoding RNAs pp Cite as Annotation of FullLength Long Noncoding RNAs with Capture LongRead Sequencing (CLS)Lnc2Cancer v updated database of experimentally supported long noncoding RNAs in human cancers Nucleic Acids Res 19 Jan 8;47(D1)D1028D1033 doi /nar/gky109646 D308D314 View in Article



Lncrnakb A Comprehensive Knowledgebase Of Long Non Coding Rnas Biorxiv



Lncexpdb Help
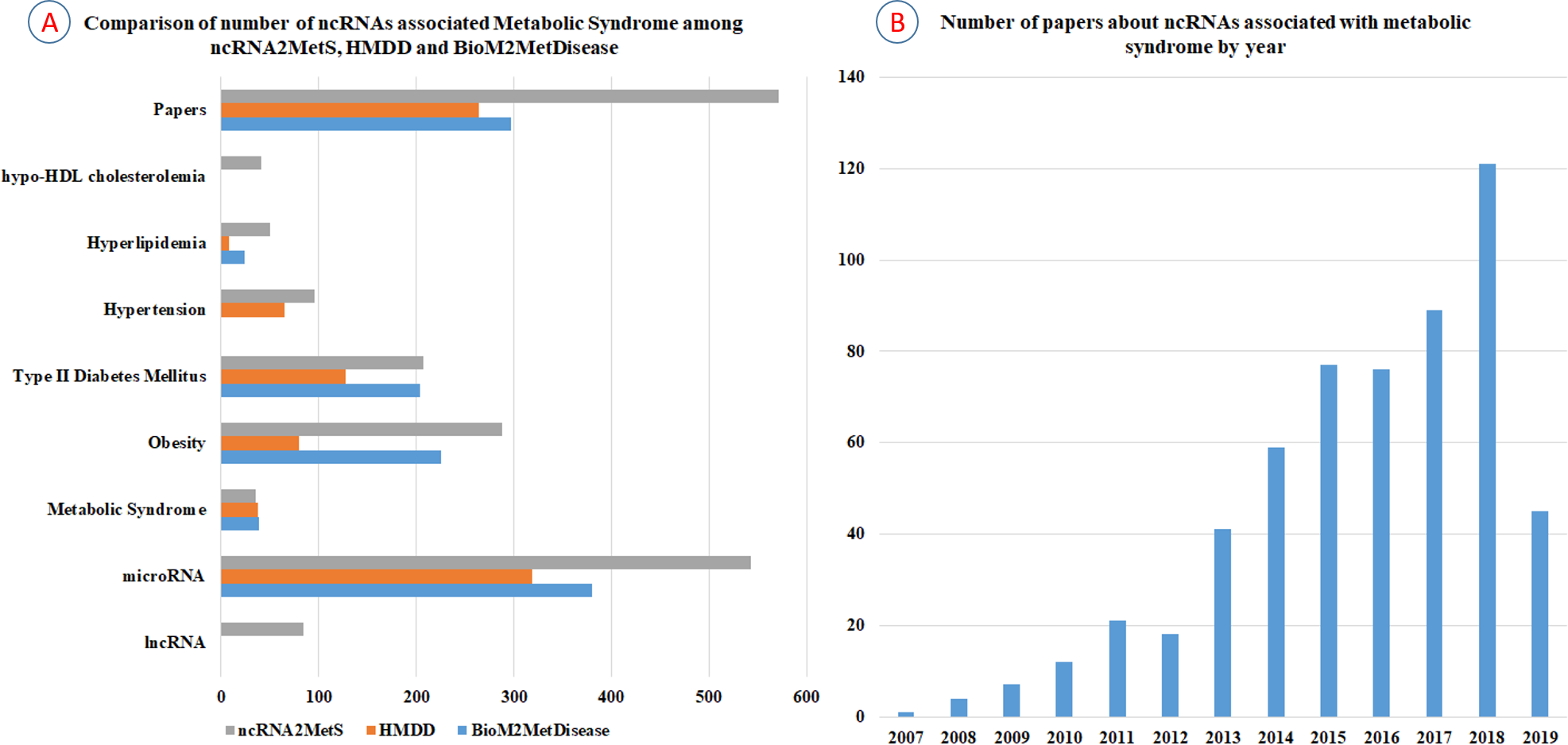


Ncrna2mets A Manually Curated Database For Non Coding Rnas Associated With Metabolic Syndrome Peerj
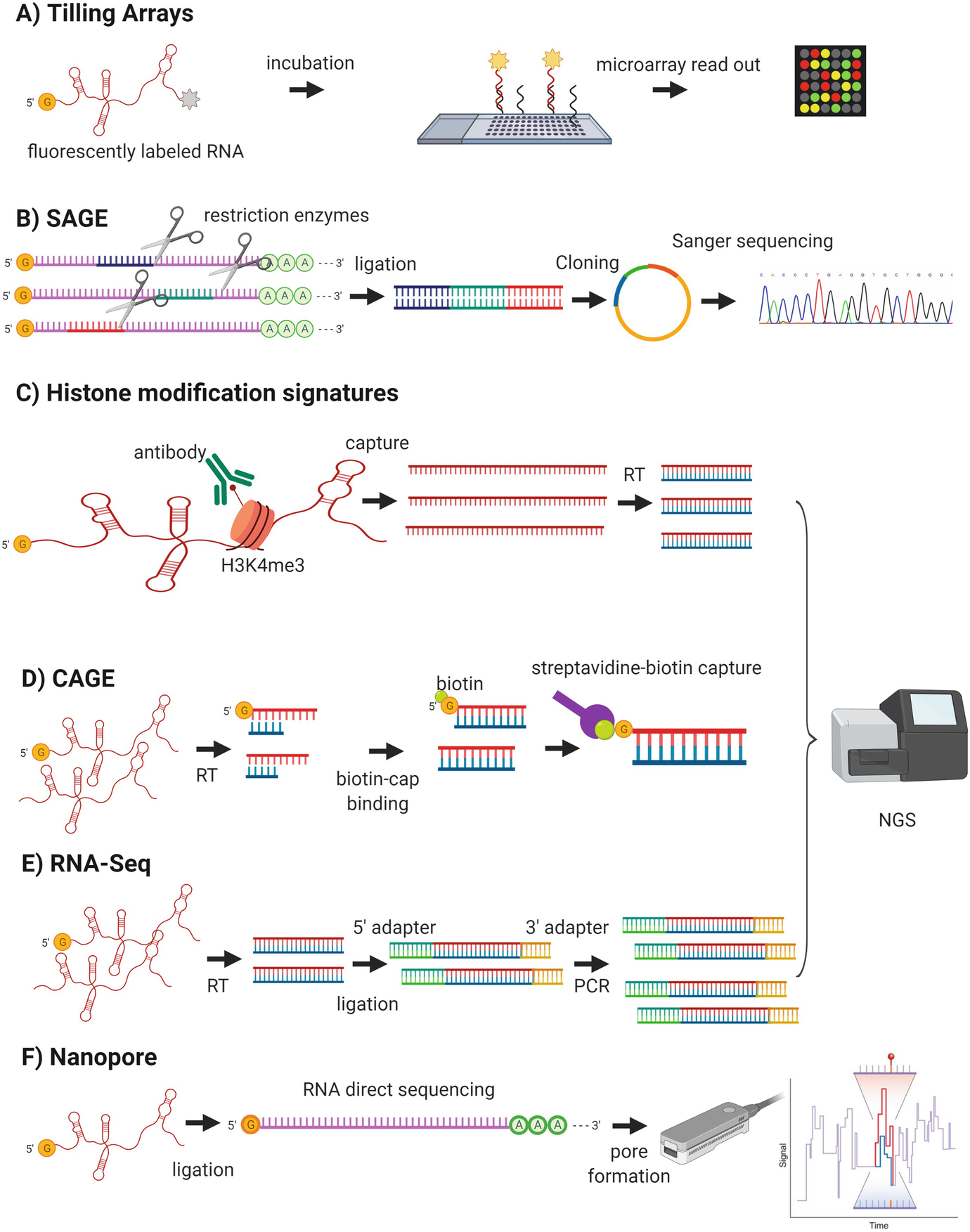


Long Non Coding Rnas Diversity In Form And Function From Microbes To Humans Springerlink



Pdf Genome Wide Identification Of Tissue Specific Long Non Coding Rna In Three Farm Animal Species
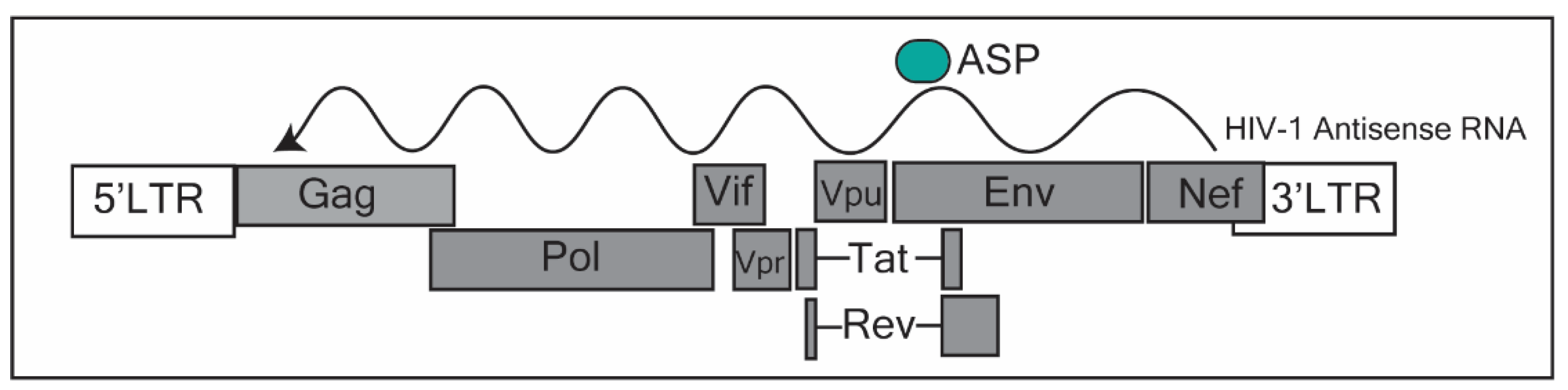


Ncrna Free Full Text Long Non Coding Rnas Mechanisms Of Action In Hiv 1 Modulation And The Identification Of Novel Therapeutic Targets Html


Frontiers Disease Causing Mutations And Rearrangements In Long Non Coding Rna Gene Loci Genetics



Lncar A Comprehensive Resource For Lncrnas From Cancer Arrays Cancer Research



The How And Why Of Lncrna Function An Innate Immune Perspective Sciencedirect



Rna Protein Interactions Disorder Moonlighting And Junk Contribute To Eukaryotic Complexity Open Biology


The Role Of Long Non Coding Rnas In Cardiac Development And Disease


Long Non Coding Rnas In Hematological Malignancies Translating Basic Techniques Into Diagnostic And Therapeutic Strategies Journal Of Hematology Oncology Full Text
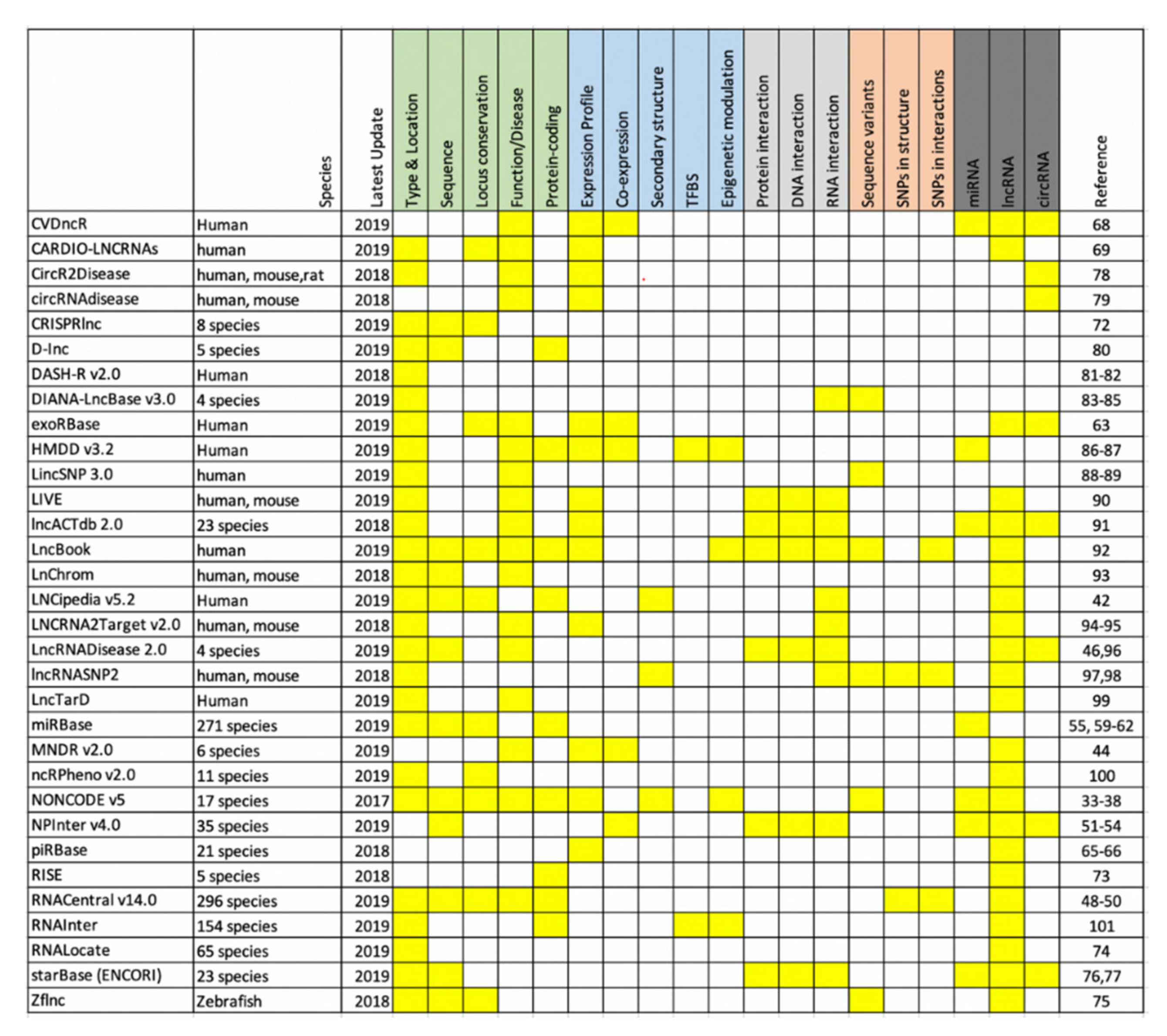


Ncrna Free Full Text Non Coding Rna Databases In Cardiovascular Research Html
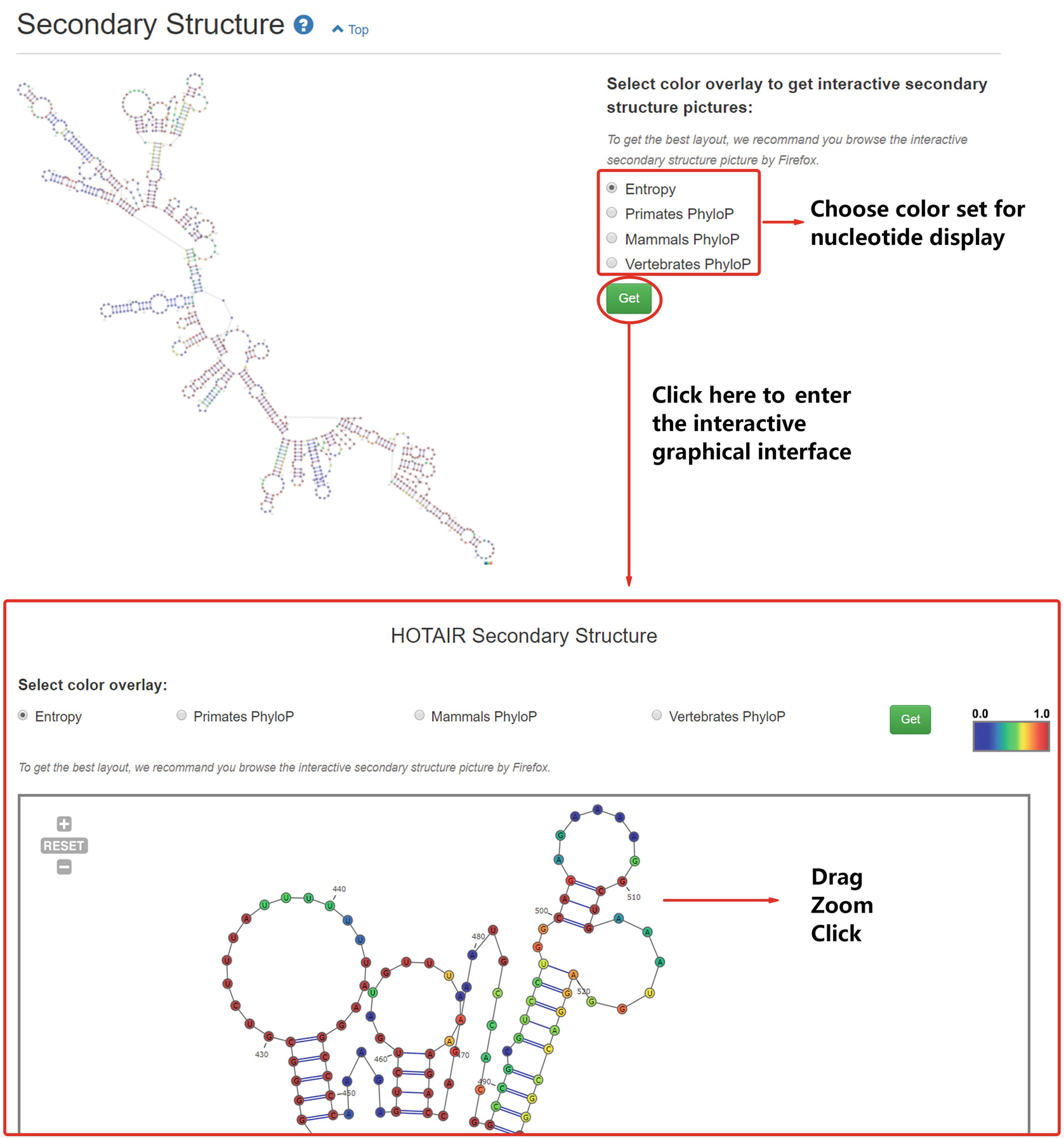


Annolnc A One Stop Portal To Systematically Annotate Novel Human Long Noncoding Rnas Springerlink



Lncar A Comprehensive Resource For Lncrnas From Cancer Arrays Cancer Research



Lncrnakb A Comprehensive Knowledgebase Of Long Non Coding Rnas Biorxiv
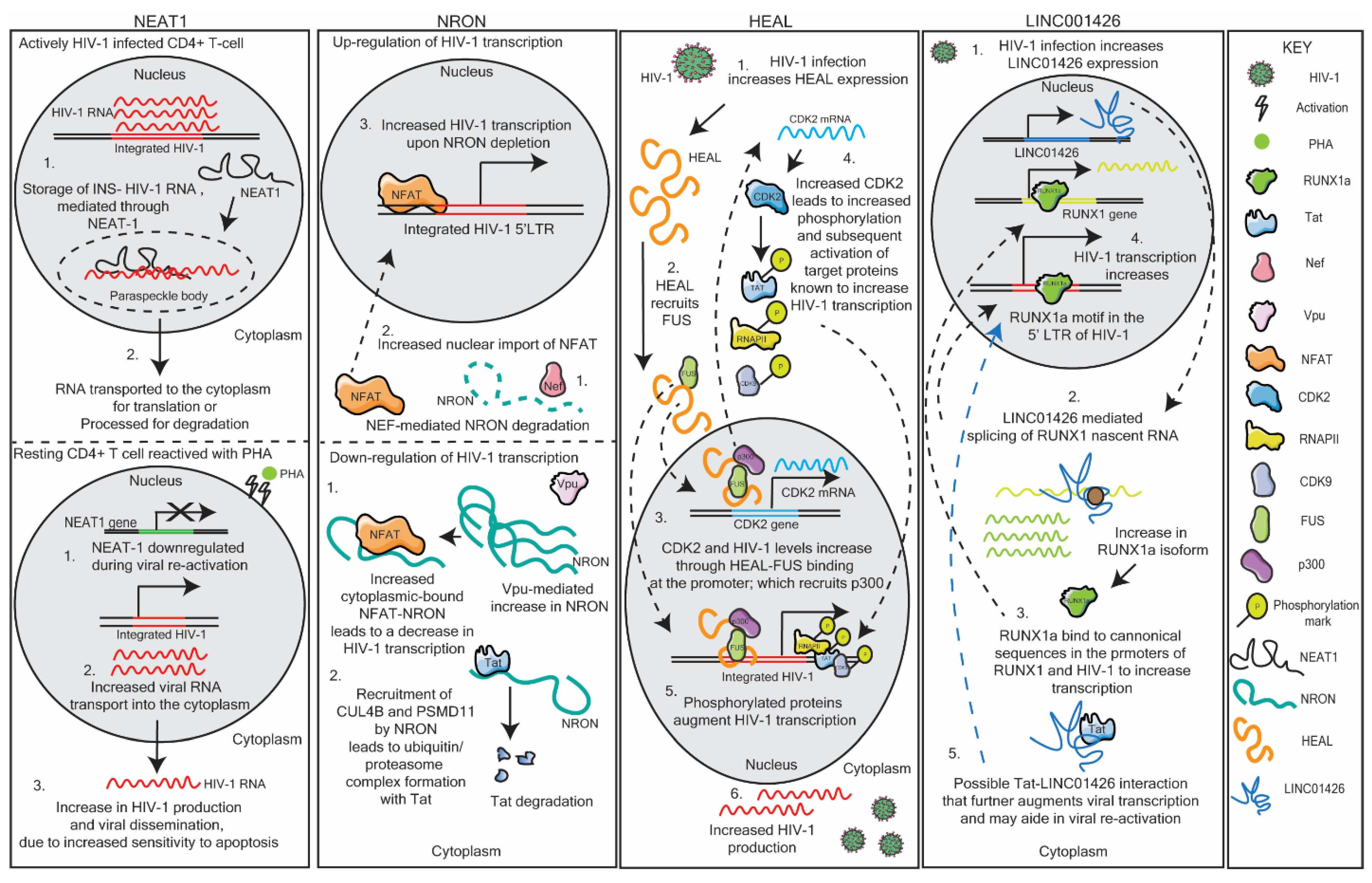


Ncrna Free Full Text Long Non Coding Rnas Mechanisms Of Action In Hiv 1 Modulation And The Identification Of Novel Therapeutic Targets Html
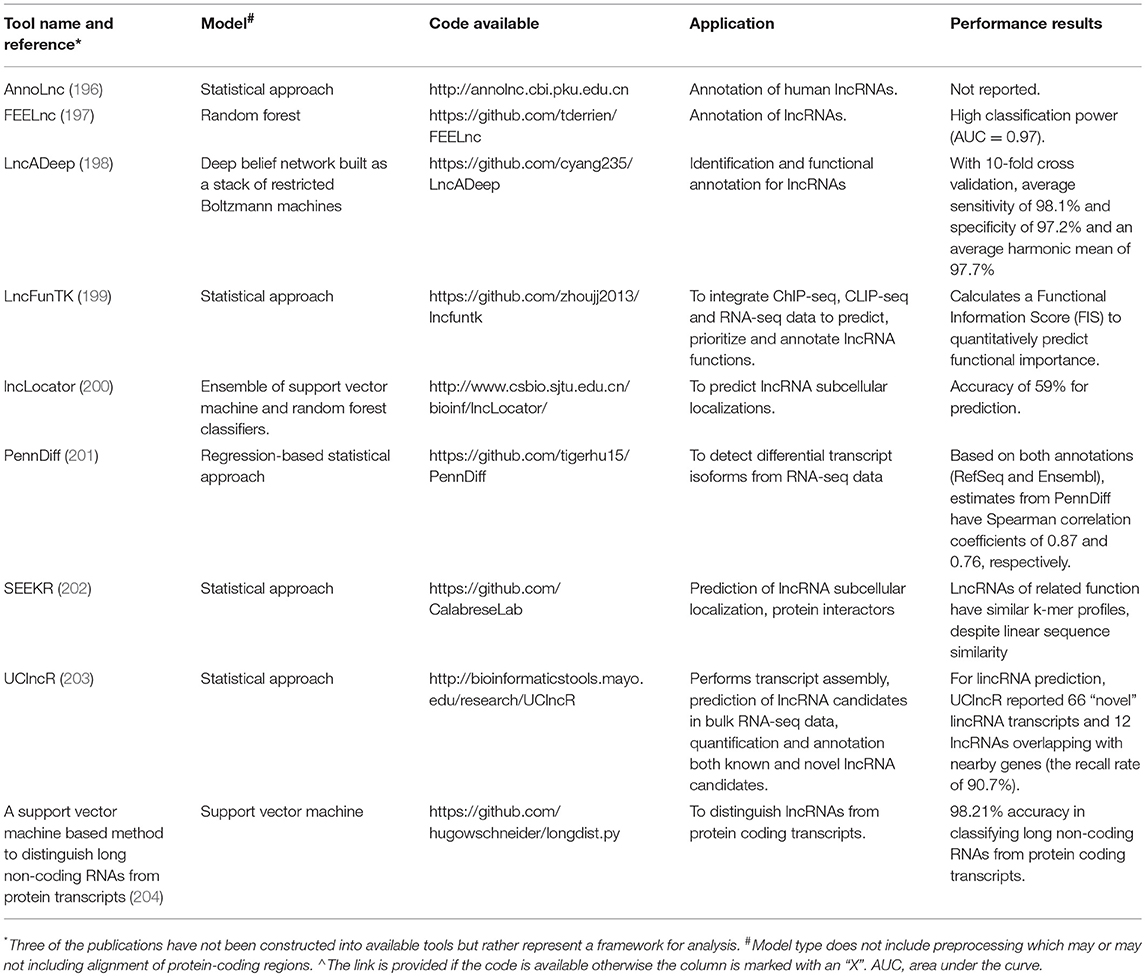


Frontiers Multi Omics Approaches To Study Long Non Coding Rna Function In Atherosclerosis Cardiovascular Medicine
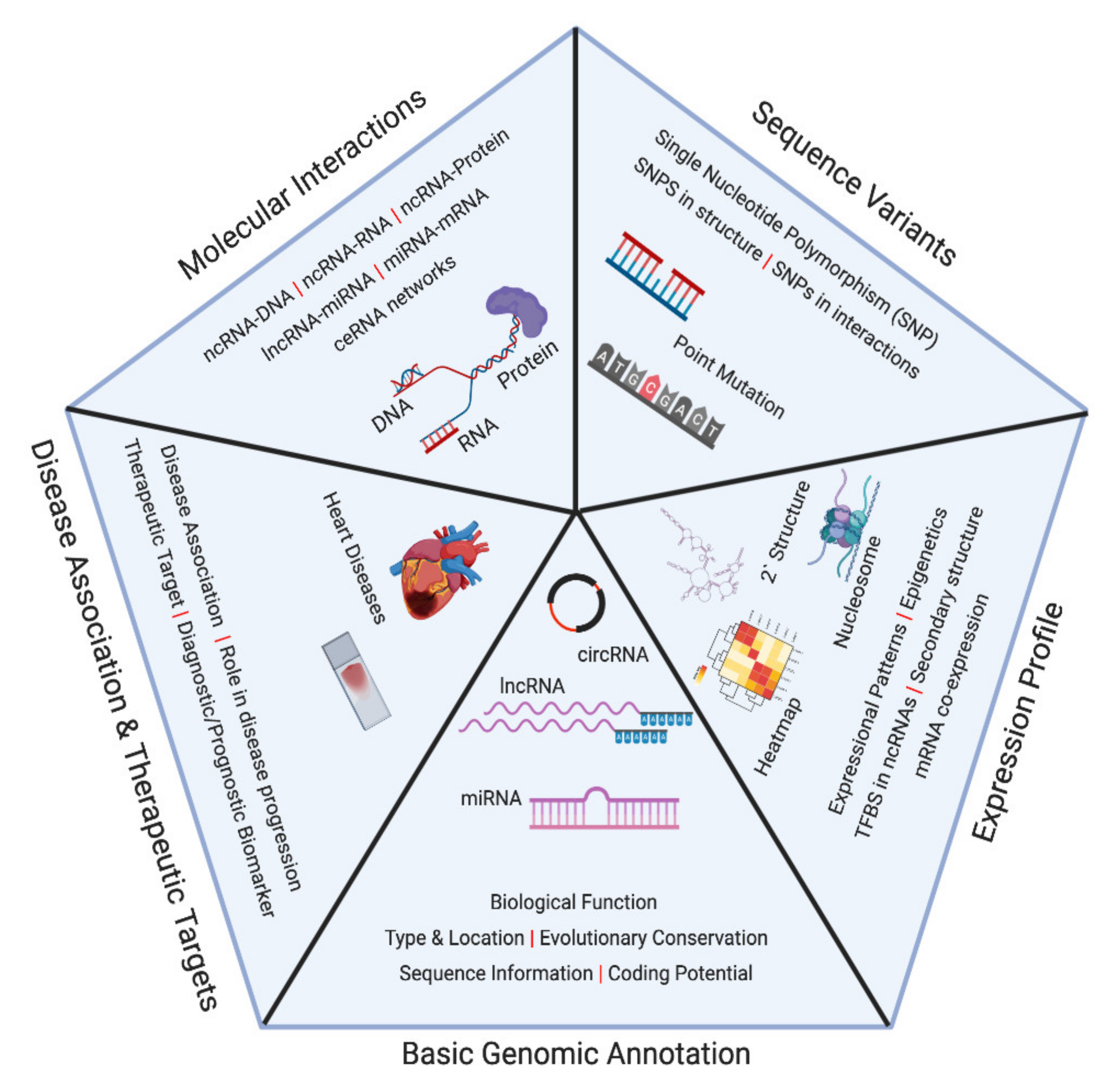


Ncrna Free Full Text Non Coding Rna Databases In Cardiovascular Research Html
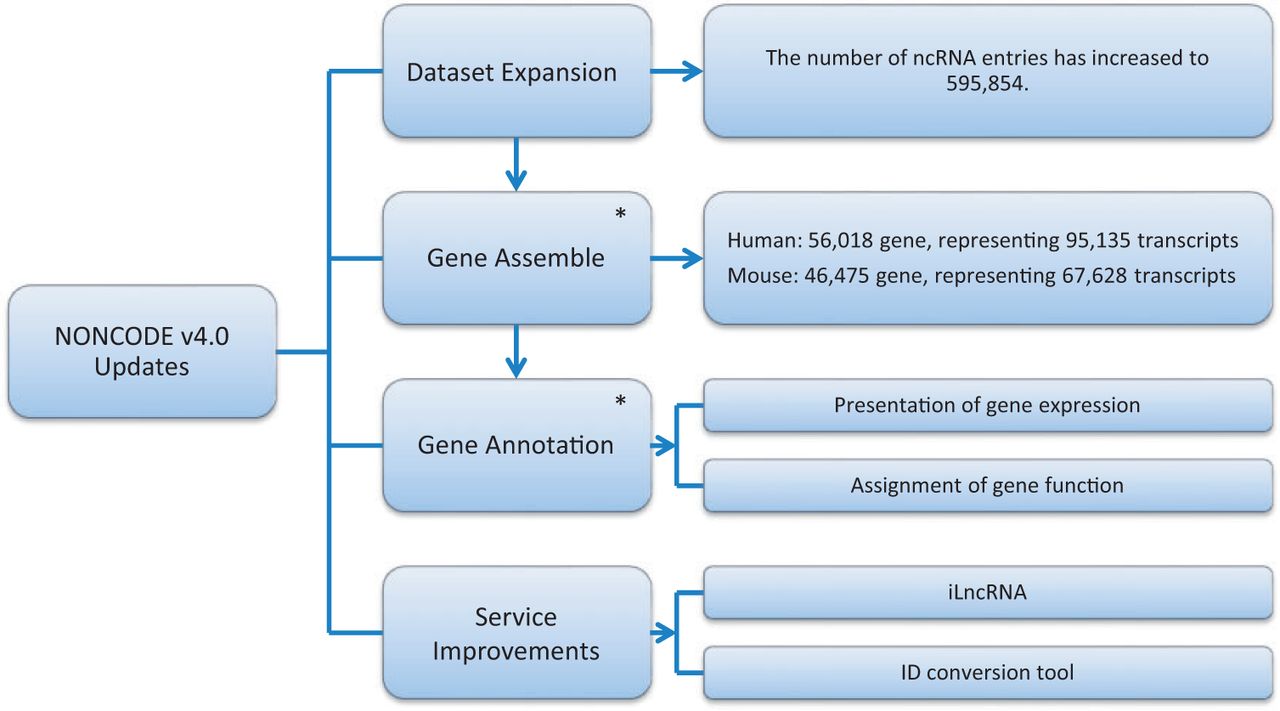


Noncodev4 Exploring The World Of Long Non Coding Rna Genes Lncrna Blog



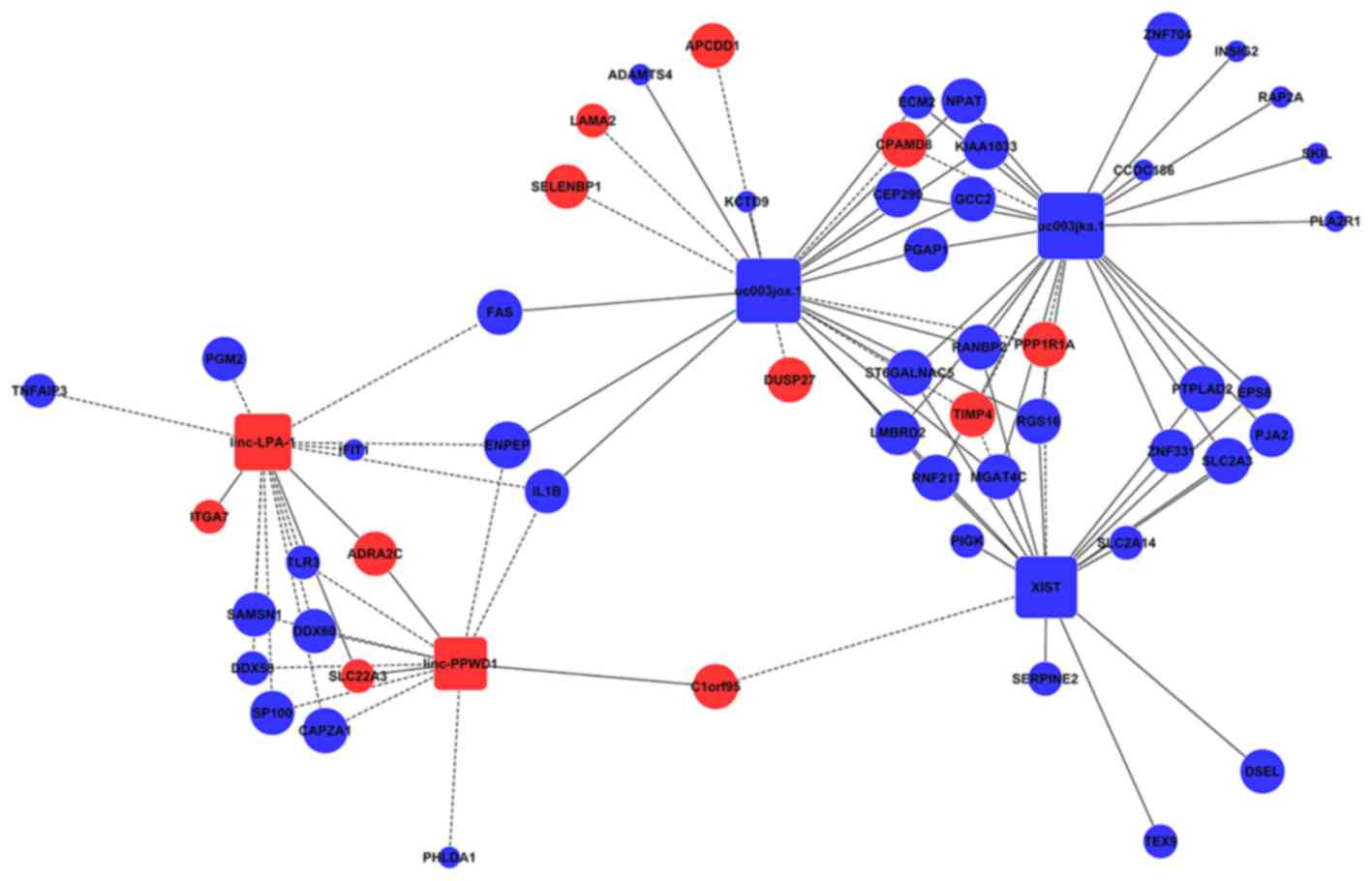
Figure 1 From Lincsnp 2 0 An Updated Database For Linking Disease Associated Snps To Human Long Non Coding Rnas And Their Tfbss Semantic Scholar
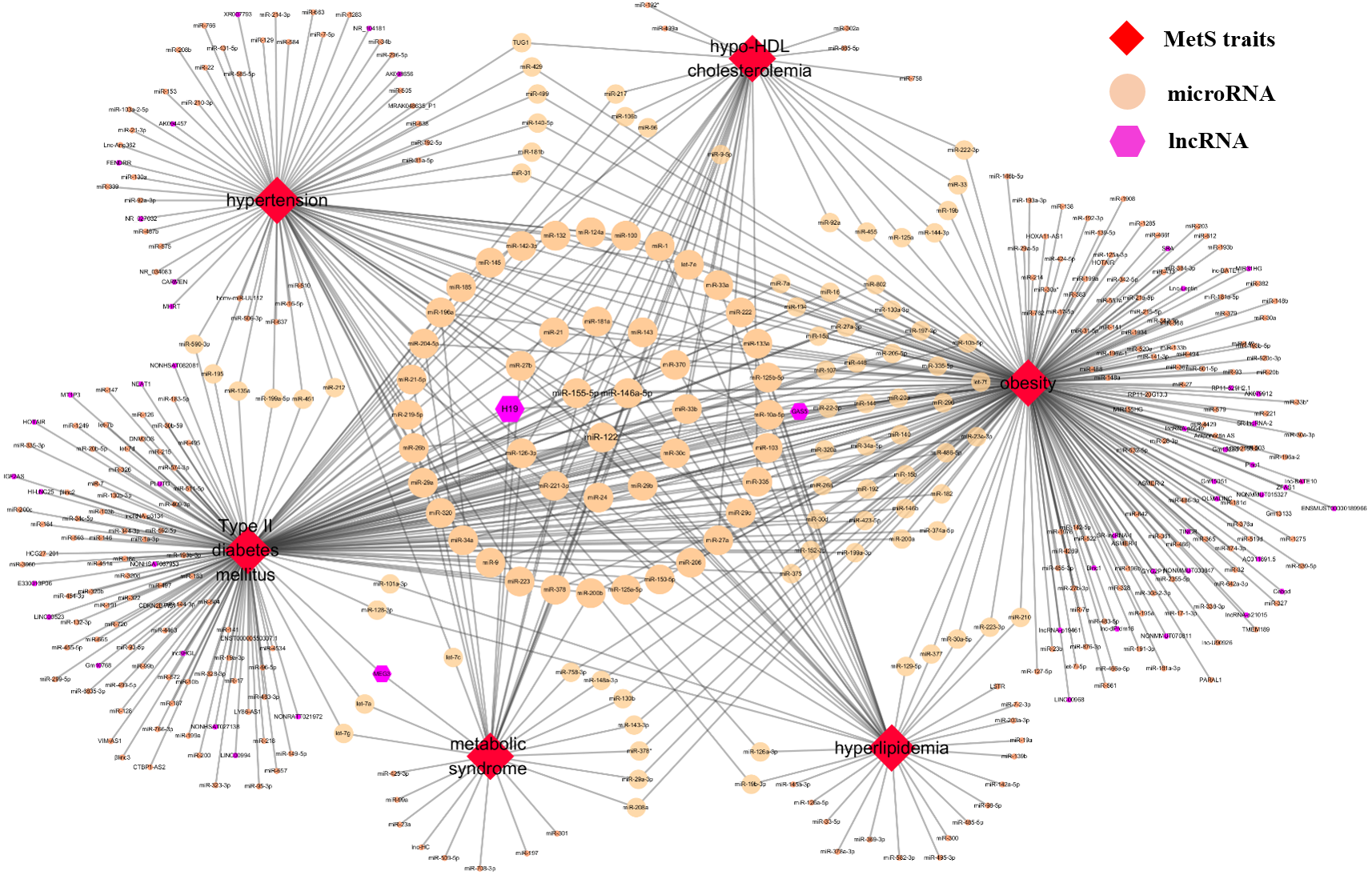


Ncrna2mets A Manually Curated Database For Non Coding Rnas Associated With Metabolic Syndrome Peerj



Lncmap Pan Cancer Atlas Of Long Noncoding Rna Mediated Transcriptional Network Perturbations Lncrna Blog



Lncexpdb Help
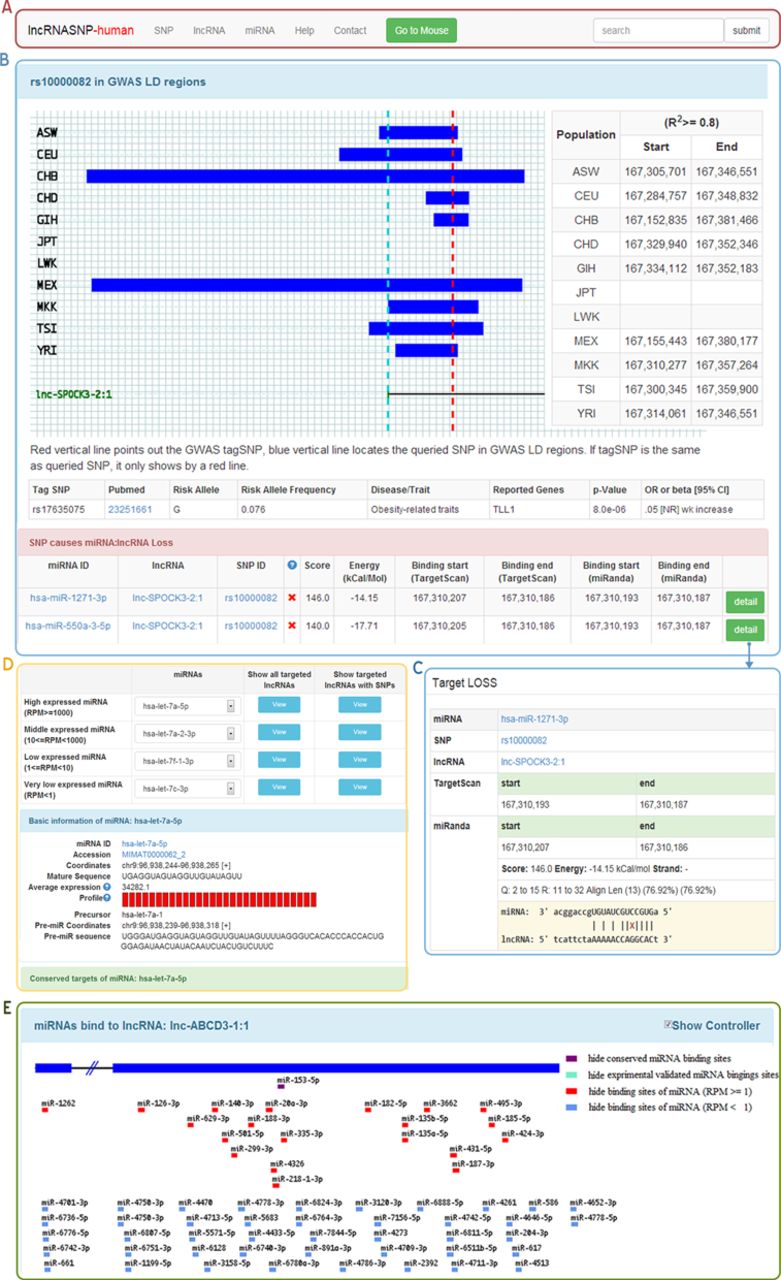


Lncrnasnp A Database Of Snps In Lncrnas And Their Potential Functions Lncrna Blog



Lncrnakb A Comprehensive Knowledgebase Of Long Non Coding Rnas Biorxiv



Non Coding Rnas Long Non Coding Rnas And Micrornas In Endocrine Related Cancers In Endocrine Related Cancer Volume 25 Issue 4 18



0 件のコメント:
コメントを投稿